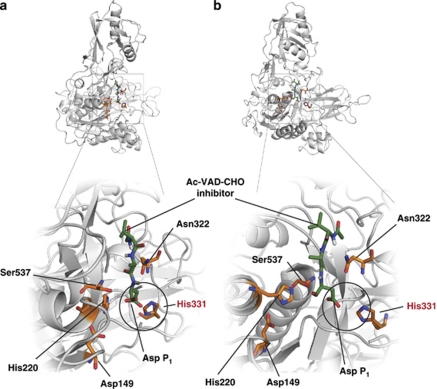
Figure 4.
A model of the phytaspase active site. A homology model of processed tobacco phytaspase based on the structure of tomato SBT3 subtilase15 (PDB 3IS6) was built using MODELLER 9.7.56 Docking calculations for Ac-VAD-CHO phytaspase inhibitor were performed using DockingServer.57 The lowest free energy of binding model was visualized using PyMOL.58 Catalytic amino-acid residues Asp149, His220, Asn322 and Ser537 are shown in orange, with O atoms in red and N atoms in blue. The same designations are employed for His331 predicted to fix the β-carboxylic group of the ligand P1 Asp through polar interactions (circled). Ligand position (green) is shown from different angles in (a) and (b)