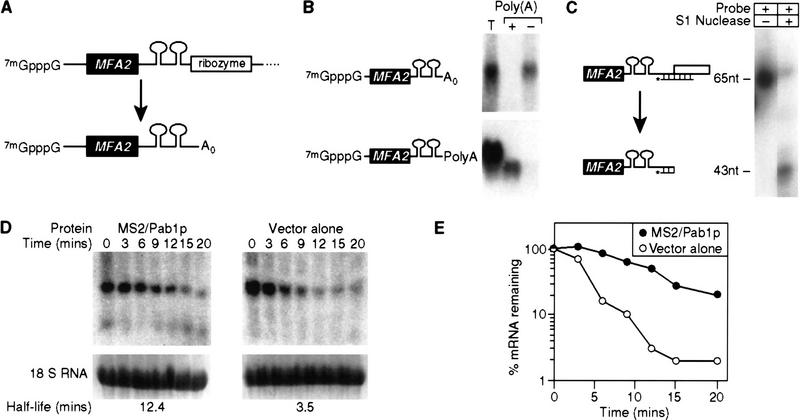
Figure 4.
Tethered Pab1p stabilizes mRNAs that do not receive a tail. (A) Strategy. The HDV self-cleaving ribozyme was inserted into the 3′ UTR of MFA2/MS2 RNA, yielding an mRNA without a poly(A) tail (A0) in vivo. (B) Oligo(dT) cellulose chromatography. RNAs were extracted from cells carrying the ribozyme-containing construct depicted in A, or the control plasmid used in previous figures. RNAs were fractionated by oligo(dT) cellulose chromatography, then analyzed by Northern blotting. The ribozyme-containing RNA is not retained by the column, whereas the control RNA is retained. (T) Total RNA prior to fractionation; (+) retained by the column; (−) not retained by the column. The mRNA runs slightly faster in the oligo(dT) retained (+) sample than in the total (T) RNA, because less RNA is loaded per lane. (C) S1 nuclease analysis. The 3′ end of mRNA prepared from a strain carrying the ribozyme-containing RNA was determined by S1 nuclease mapping. The position of undigested probe (65 nucleotides) and of the protected species (43 nucleotides) are shown. The prominent 3′ terminus lies at the expected position for ribozyme cleavage. (D) Decay of ribozyme-cleaved RNA by a transcriptional pulse-chase experiment and Northern blotting. The turnover rate of ribozyme-cleaved reporter mRNAs was determined in strains with (left) or without (right) MS2/Pab1p. Time (in min) following transcriptional repression are given directly at top; half-lives are at bottom. (E) Quantitation of results. Amounts of mRNA were normalized to the level of 18S rRNA, at bottom of each lane in D. (•) MS2/Pab1p; (○) vector alone.