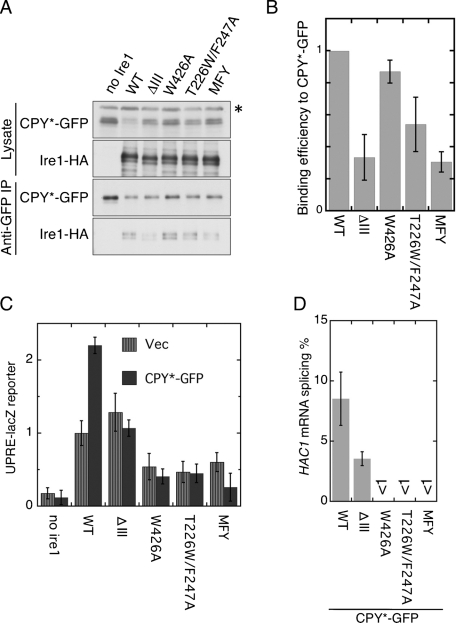
FIGURE 4:
Effect of the CSSR mutations on relationship between Ire1 and CPY*-GFP. (A) The ire1Δ strain KMY1015 transformed with both pRS313-GAL1pr-CPY*-GFP and pRS426-IRE1-HA or its mutants (or empty vector pRS426; no ire1) was cultured in galactose-containing medium and analyzed as per Figure 3A. The asterisk denotes nonspecific bands. (B) The ratios of Ire1-HA to CPY*-GFP signal in anti-GFP IP in triplicate experiments demonstrated in (A) were normalized to that of wild-type Ire1 and are presented as “Binding efficiency to CPY*-GFP.” (C) The ire1Δ strain triply transformed with UPRE-lacZ plasmid pCZY1, plasmid pRS313-GAL1pr-CPY*-GFP (or empty vector pRS313; Vector), and pRS315-IRE1-HA (WT) or its mutants (or empty vector pRS315; no ire1) was cultured in galactose-containing medium before measuring cellular β-galactosidase activity, the values of which are normalized against that of WT IRE1 Vector cells (set at 1.00). Error bars represent the SDs from three independent transformants. According to Student's t test, the values of all CPY*-GFP mutant Ire1 samples are statistically different from those of the CPY*-GFP WT IRE1 sample (p < 0.01). Also, when not carrying the CPY*-GFP plasmid, the WT IRE1 sample exhibited statistical difference from the W426A, T226W/F247A, and MFY mutant samples (p < 0.05). (D) Cells used in (C) were analyzed by RT-PCR to evaluate splicing efficiency of the HAC1 mRNA.