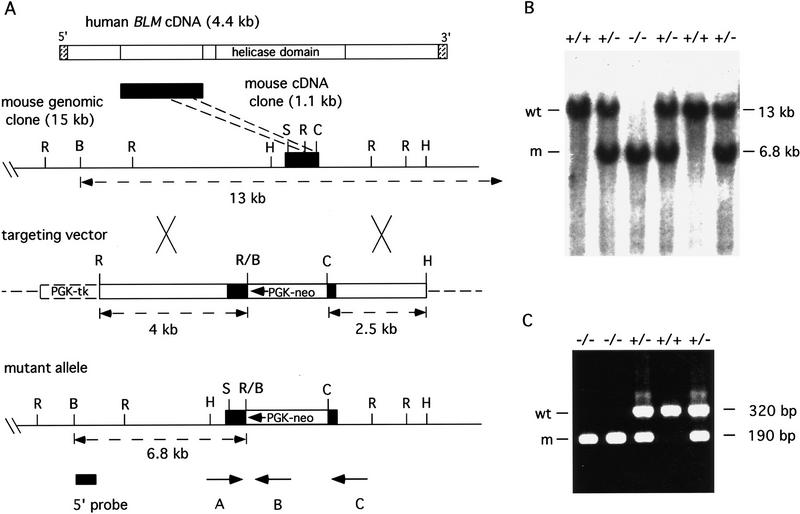
Figure 1.
Targeted disruption of the mouse Blm gene. (A) Shown is the alignment of the mouse Blm cDNA fragment upstream of the helicase domain present in the human BLM coding sequence. The map of mouse Blm genomic clone DNA is shown with restriction sites and the targeted coding region indicated by black shading. (B) BamHI; (C) ClaI; (H) HindIII; (R) EcoRI; (S) StuI. The targeting vector contained the neo resistance and the herpes simplex virus tk genes under the control of the mouse PGK promoter. Homologous recombination resulted in deletion of 180 bp of sequence between the EcoRI and ClaI sites and placed the direction of transcription of PGK–neo opposite that of Blm in the mutant allele. A Blm gene 5′-flanking probe used for screening ES cell clones and mice is indicated; also the predicted sizes of wild-type and mutant fragments that resulted from restriction digestion of genomic DNA with BamHI are shown. Positions of oligonucleotide primers used for PCR genotype analysis are indicated (A, B, and C). (B) Southern blot analysis of BamHI-digested embryo genomic DNA. Fragments that hybridized to the 5′ probe are 13 kb for the wild-type allele (wt) and 6.8 kb for the mutant allele (m). (C) PCR analysis of embryo yolk sac genomic DNA. With the simultaneous addition of three oligonucleotide primers to the PCR, all possible genotypes can be obtained. Reaction products that resulted from primer pair AB; 190-bp mutant allele (m), and primer pair AC; 320-bp wild-type allele (wt), are visualized by agarose gel electrophoresis and ethidium bromide staining. Genotypes are shown at top for each assay in B and C.