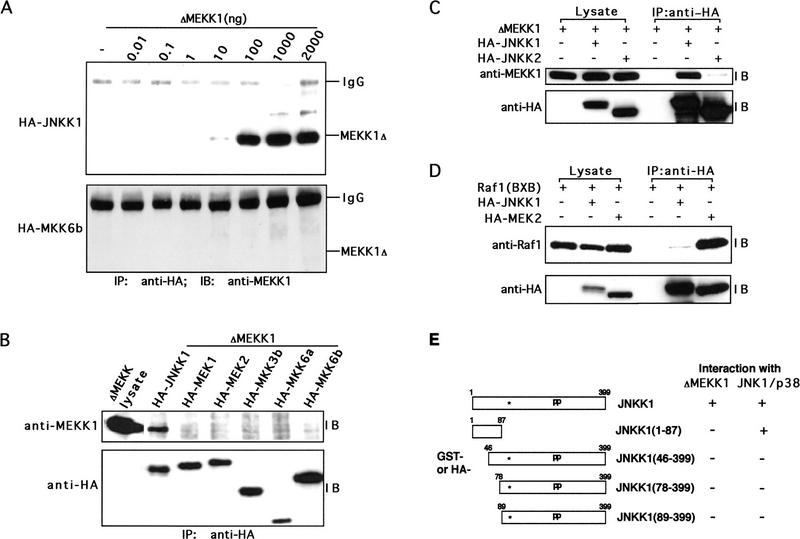
Figure 3.
JNKK1 specifically interacts with MEKK1. (A) JNKK1, but not MKK6, forms stable complexes with MEKK1. Lysates of Cos-1 cells cotransfected with the indicated amounts of ΔMEKK1 and either HA–JNKK1 or HA–MKK6b expression vectors were immunoprecipitated (IP) with anti-HA. The precipitates were separated by SDS-PAGE and analyzed by immunoblotting (IB) with anti-MEKK1. The efficiencies of HA–JNKK1 and HA–MKK6 expression were similar (data not shown). (B) JNKK1, but not other MAPKKs, interacts with MEKK1. Lysates of Cos-1 cells transiently expressing HA–MAPKKs were mixed with lysates containing ΔMEKK1. The mixtures were immunoprecipitated with anti-HA, the immunocomplexes were separated by SDS-PAGE and immunoblotted sequentially with anti-MEKK1 (top) and anti-HA (bottom). (C) JNKK1, but not JNKK2, interacts with MEKK1. Cos-1 cells were transiently transfected with ΔMEKK1 and either HA–JNKK1, HA–JNKK2, or empty expression vector. Cell lysates were either directly separated by SDS-PAGE or immunoprecipitated with anti-HA as indicated and then resolved by SDS-PAGE. Immunoblot analyses were performed with anti-MEKK1 (top) or anti-HA (bottom). (D) Raf-1 interacts with MEK2 but not with JNKK1. Cos-1 cells were transfected with Raf1(BXB), HA–JNKK1, and HA–MEK2 expression vectors, as indicated. Cell lysates were analyzed as described in A. Protein expression was evaluated by separating one-fortieth of each lysate directly by SDS-PAGE and immunoblotting with anti-Raf-1 (top) or anti-HA (bottom) antibodies. The same antibodies were used for immunoblotting of anti-HA immunoprecipitates. (E) Amino-terminal extension of JNKK1 is required for interacting with MEKK1 and JNKs(p38). Schematic representation of JNKK1 deletion mutants with either GST or HA tags at their amino termini. The location of the ATP-binding site is indicated by an asterisk and the activating phosphoacceptor sites by pp. GST-tagged full-length and truncated JNKK1 proteins were expressed either in Cos-1 cells or in bacteria and their binding to ΔMEKK1, JNK, and p38 was examined by in-vitro mixing–coprecipitation assays, as described in B. The results are indicated on the right.