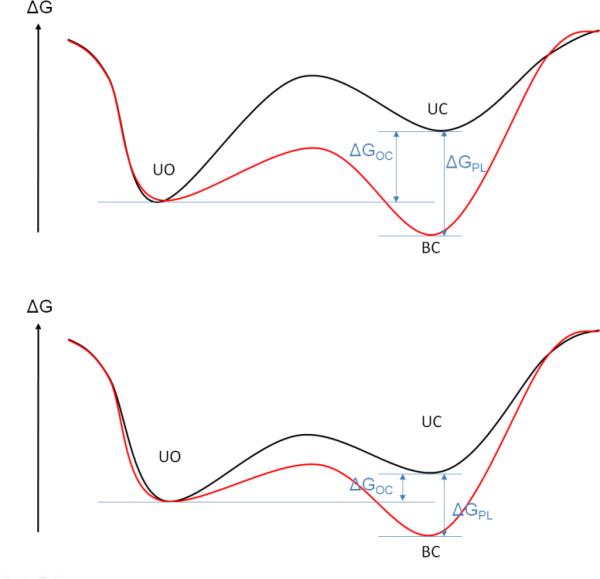
Figure 3.
Two different free-energy models that characterize protein-ligand systems, likely representing the induced-fit (top panel) and the conformational-selection (bottom panel) mechanisms of binding are shown. The black curve models the free energy of the unbound form of the protein as a function of an arbitrary reaction coordinate that describes the conformational change between the open and closed form of the protein. The red curve displays the associated model of the free energy of the bound form of the protein. ΔGOC is the free-energy difference between the open and closed state of the ligand-free protein, and ΔGPL is the stabilizing free energy of the protein upon ligand binding. The free energy ΔGPL contains many contributions including the direct protein-ligand interactions typically affected by water screening, hydrophobic association, and the conformational and vibrational entropy of protein and ligand.