Figure 6.
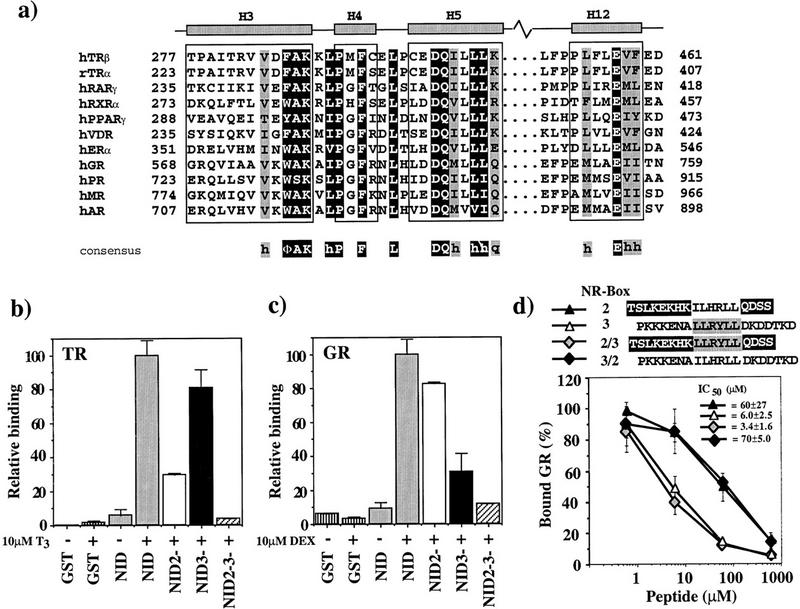
(a) Sequence alignment of LBD helices 3, 4, 5, and 12. NR sequences (Wurtz et al. 1996) are shown for LBD regions that interact with GRIP1. Residues representing the NR signature sequence, ΦAKxhPxFxxLxxxDQxxhh, are on dark background. Conserved residues, always hydrophobic (h) or strong polar (q), are shaded. The borders for helices H3, H4, H5, and H12 are those for the TRβ LBD. (Φ) Bulky hydrophobic residue. (b,c) GR and TR interact preferentially with different GRIP1 NR boxes. Labeled TRβ (b) or rat GR (c) (10 nm) was incubated with 1.6 μm glutathione–agarose-bound GST (vertically striped), GST–NID (shaded) or the NID variants NID2− (open), NID3− (solid), and NID2−3− (hatched) in the absence (−) or presence (+) of 10 μm T3 (TR) or 10 μm dexamethasone (DEX) (GR). The amount of bound receptor is normalized to the fraction bound to GST–NID (100%). The data represent the average and standard deviation of about three independent experiments. (d) Preferential interaction of GR with NR-box 3 is not specified by sequences adjacent to the LLRYLL motif. Labeled GR (10 nm) was incubated with 1.6 μm glutathione–agarose-bound GST–NID3− in the presence of 10 μm DEX and increasing concentrations of NR-box 2 peptide (solid triangle), NR-box 3 peptide (open triangle), and either a chimeric peptide bearing the LLRYLL motif of NR-box 3 with the adjacent sequences from NR-box 2 (shaded diamond), or a chimeric peptide bearing the ILHRLL motif of NR-box 2 with the adjacent sequences from NR-box 3 (solid diamond). The amount of bound receptor is relative to the amount of retained receptor in the absence of peptide. The data and IC50 values represent the average and standard deviation of three independent experiments.
