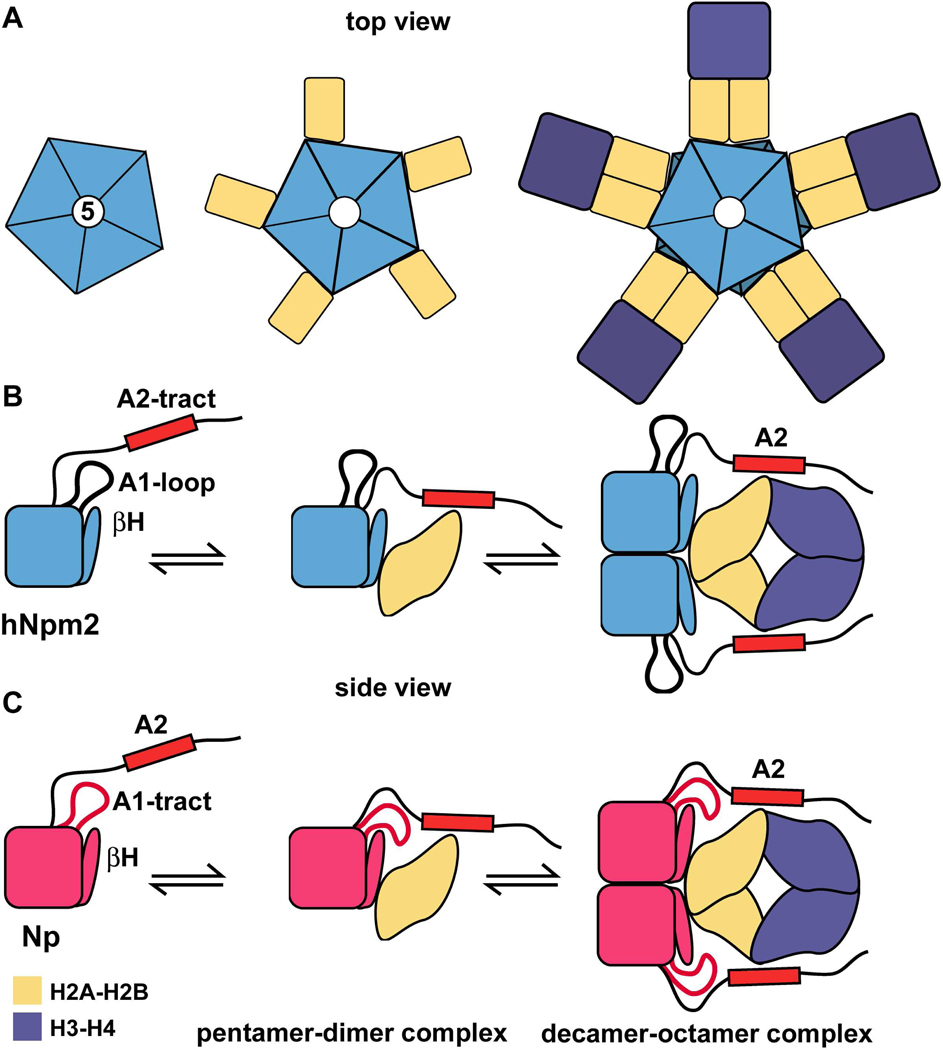
Figure 7. Models of histone binding by human and Xenopus nucleoplasmin.
(A) Nucleoplasmin pentamers may bind histone dimers on their lateral surfaces. Subsequent binding of H3-H4 tetramers would form a bridge between two pentamer-histone dimer complexes.
(B) A hypothetical, stepwise assembly of an Npm2 decamer/core histone complex is shown in a side view. This occurs when Npm2 pentamers bind a mixture of H2A-H2B dimers (gold) and H3-H4 tetramers (purple). The C-terminal A2-tract is required to form a stable histone complex and is shown schematically as a red rectangle, while the A1-loop (thick black line) is not required. For simplicity only 1 subunit in each Npm2 pentamer is shown and additional domains in the C-terminal tail are not shown in this figure. (C) Np decamers create a central hub when they bind a mixture of H2A-H2B dimers (gold) and H3-H4 tetramers to form histone octamers on the chaperone. Both the A1-tract (in red) and A2-tracts (red rectangle) help stabilize the complex.