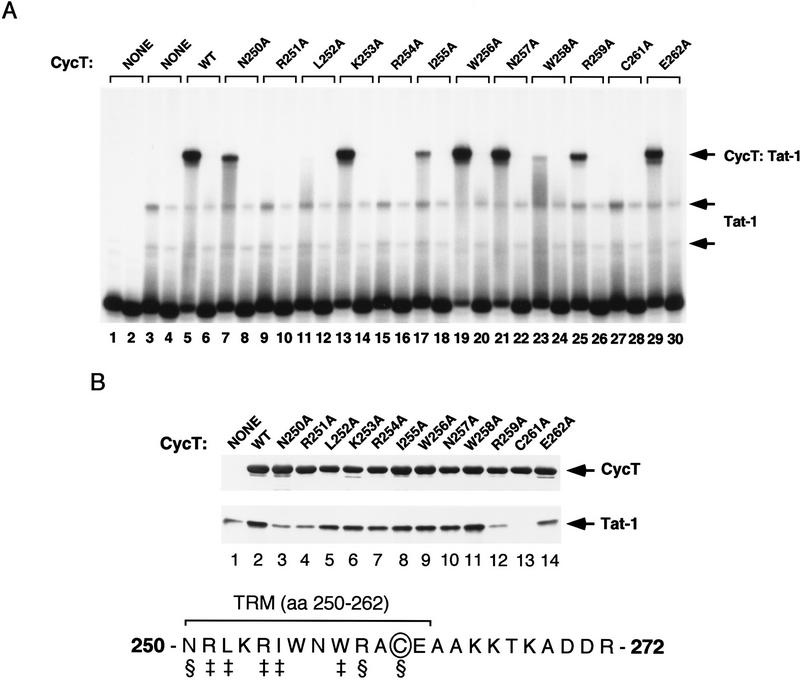
Figure 3.
Alanine-scanning mutagenesis of the TRM of hCycT1. (A) Analysis of the ability of different alanine-substituted hCycT1 (amino acids 1–303) proteins to enhance the binding of HIV-1 Tat to TAR RNA in a gel-mobility shift experiment. Reactions contained either wild-type TAR RNA (odd-numbered lanes) or TAR loop mutant (+29/+34) RNAs (even-numbered lanes). Reactions included 175 ng (GST-cleaved) of Tat-1 (lanes 3–30) and 40 ng (GST-cleaved) of hCycT1 (amino acids 1–303). The position of each mutation is indicated above each lane. (WT) Wild-type hCycT1 (amino acids 1–303). (B) Analysis of the ability of the different hCycT1 mutants to interact directly with HIV-1 Tat in vitro. Reactions included 60 ng (GST-cleaved) of HIV-1 Tat and 250 ng of GST–hCycT1 (amino acids 1–303), either wild-type (WT) or mutant, as indicated above each lane. The hCycT1 protein was visualized with a monoclonal antibody to GST, and the (GST-cleaved, HA-tagged) Tat was visualized with an anti-HA monoclonal antibody. Lane 1 represents 10% of the input protein (200 ng) of the wild-type Tat protein. The TRM within hCycT1 is shown at the bottom. The residues designated with the ‡ symbol are required for TAR RNA recognition, whereas the residues indicated with the § symbol are needed to bind Tat.