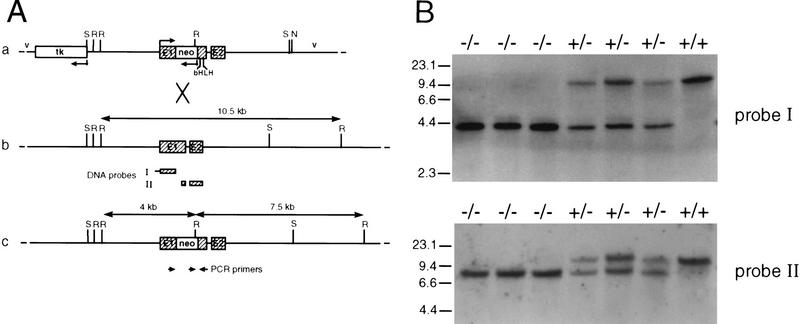
Figure 1.
Targeted inactivation of the p48 gene by homologous recombination and genotype analysis of heterozygote interbreeding. (A) Schematic maps of the targeting vector (a), the wild-type p48 locus (b) and the mutated allele (c). Exons are shown as hatched boxes and intronic or gene flanking sequences as heavy lines. Bacterial vector sequences (v) are designated by thin lines. The direction of transcription is indicated by bent arrows. The herpesvirus tk gene and the neo gene used for negative and positive selection, respectively, are in the opposite transcriptional orientation to the p48 gene. The neo cassette was inserted into exon 1 (E1) of the p48 gene upstream of the bHLH sequence. Cleavage sites for restriction endonucleases SacI (S) and NotI (N) were those used for construction and linearization of the targeting vector, respectively. Cleavage at EcoRI sites (R) was used to distinguish between wild-type and mutant alleles. The diagnostic EcoRI fragments are highlighted by double-headed arrows, and their size is shown. The origin of radiolabeled DNA probes used for filter hybridizations in B is indicated below the map of b. The position and polarity of oligonucleotide primers used for PCR amplification are represented by the small arrows below the map of c. (B) Southern analysis of EcoRI-digested genomic DNA of newborn littermates from a +/− interbreeding. DNA probes I and II both detect a 10.5-kb restriction fragment of the wild-type allele. In addition, probe I detects a 4-kb fragment, and probe II a 7.5-kb fragment, of the mutant allele. Subsequent genotype analysis was carried out by PCR on tail DNA with the gene-specific primers shown in A (data not shown).