Figure 2.
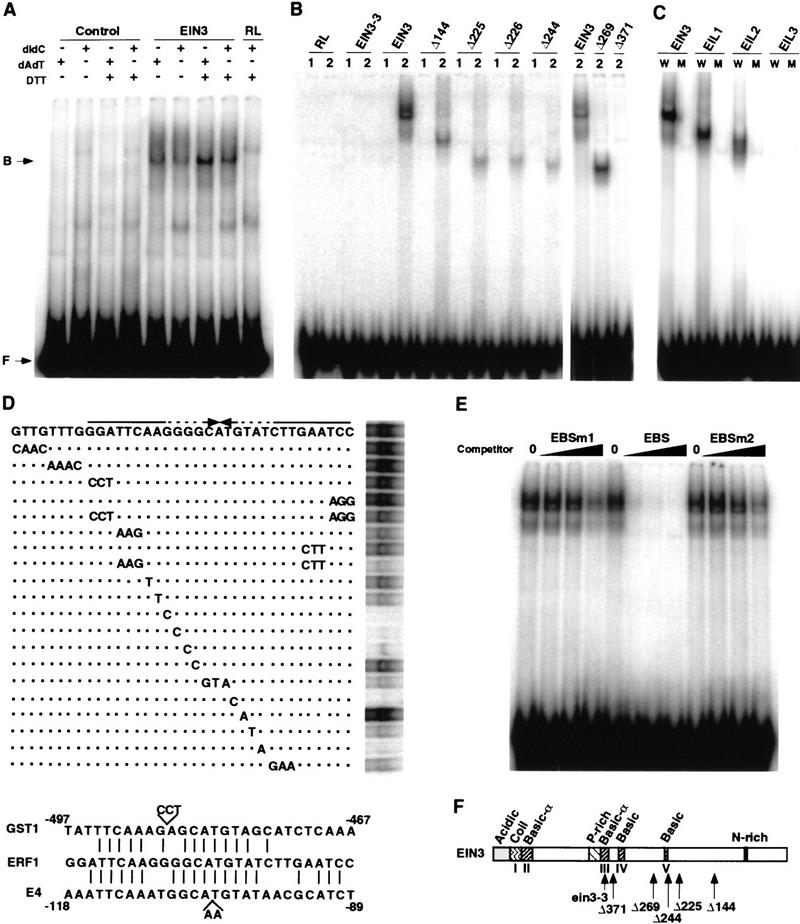
EIN3 is a sequence-specific DNA-binding protein. (A) EMSA of in vitro-translated EIN3 protein binding to the −1238 to −950 fragment of the ERF1 promoter. A control protein (control) or mock-translated reticulocyte lysates (RL) were used in the indicated lanes. (B) EMSA of EIN3-3 mutant protein, EIN3, and several carboxy-terminal deletion derivatives bound to fragments −1238 to −1204 (1) and −1213 to −1178 (2) of the ERF1 promoter. (C) Binding of EIL proteins to the EIN3 target site in the ERF1 promoter. EMSA was performed with in vitro-translated EIN3, EIL1, EIL2, and EIL3 proteins and the wild-type EIN3-binding site (W) or a mutant version (M) corresponding to the mutant G17 to C in Fig. 2D. (D; top) Scan mutagenesis of the EBS. Wild-type EBS is shown with the palindromic repeats indicated by arrows. Base changes in the mutants tested are indicated on the lines below. Dashed lines indicate mismatches. Dots indicate similar bases as in the wild-type EBS. (D; bottom) Sequence alignment of the EBS and a fragment of the promoters of the E4 and GST1 genes (including the ERE). (E) Competition of the EIN3–EBS complex formation by addition of an excess of unlabeled EBS or two mutant versions, EBSm1 and EBSm2 (see Materials and Methods). No competitor was added in the lanes labeled as 0. Black wedges represent increasing amounts of competitor (20, 60, and 200 ng). One nanogram of labeled EBS was used per lane. (F) Summary of EIN3 structural features and mutants used in EMSA experiments (adapted from Chao et al. 1997).
