Figure 1.
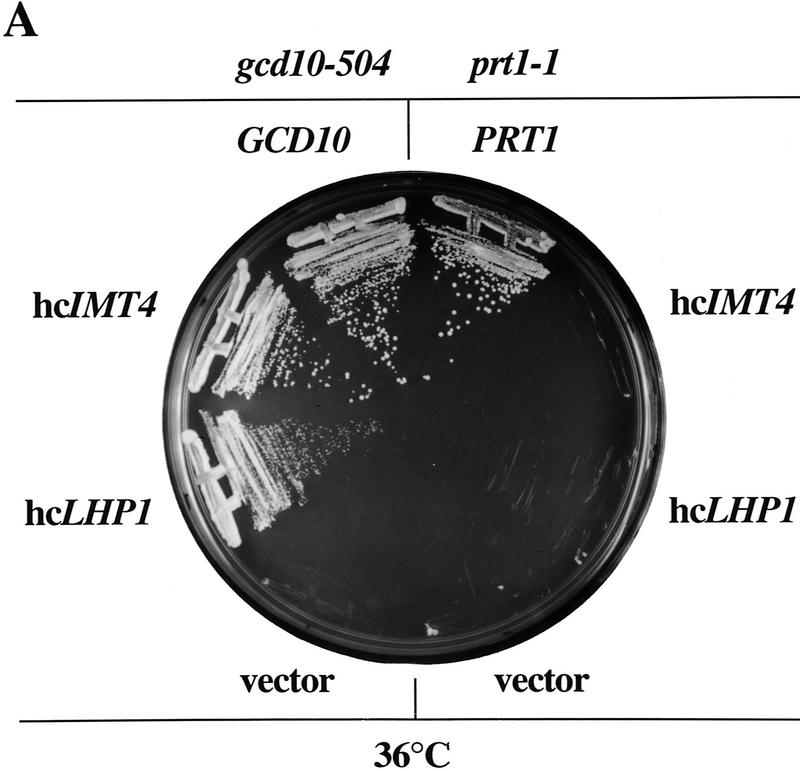
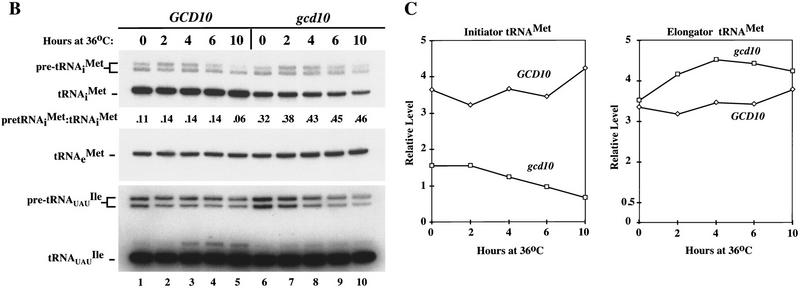
High-copy suppressors of gcd10-504 overcome a defect in accumulation of tRNAiMet (A) Transformants of strains H2457 (gcd10-504) and H1676 (prt1-1) containing low-copy plasmids bearing GCD10 (pMG107) (Garcia-Barrio et al. 1995) or PRT1 (p2625), respectively, or high-copy plasmids bearing IMT4 (pC44) (Cigan et al. 1988), LHP1 (p2626), or empty vector YEp24 were streaked for single colonies on minimally supplemented SD plates and incubated at 36°C for 2 days. (B) Northern blot analysis of total RNA (5 μg) isolated as described (Kohrer and Domdey 1991) from strain H2457 (gcd10-504) bearing GCD10 on low-copy plasmid pMG107 (GCD10 lanes) or empty vector YEp24 (gcd10 lanes) grown in supplemented SD at 26°C to mid-exponential phase (0 hr at 36°C) and shifted to 36°C for 2, 4, 6, or 10 hr. The RNAs were separated on an 8% polyacrylamide-bis-acrylamide (19:1), 8.3 m urea gel by electrophoresis and transferred to Hybond-N+ membranes (Amersham). The blot was probed using a radiolabeled oligonucleotide that hybridized specifically to tRNAiMet, stripped and reprobed with radiolabeled oligonucleotides specific for tRNAeMet or tRNAUAUIle. Direct quantitation of all hybridized probes was conducted by PhosphorImager analysis using a Storm 860 apparatus (Molecular Dynamics) and ImageQuant software. The positions of pre-tRNAiMet species, mature tRNAiMet, tRNAeMet, pre-tRNAUAUIle, and mature tRNAUAUIle are indicated at left. The relative intensities of the hybridization signals were quantified for mature tRNAiMet and pre-tRNAiMet , and the ratios of pre-tRNAiMet to mature tRNAiMet are listed under the appropriate lanes. The species that migrated just above mature tRNAUAUIle and accumulated at high temperature most likely represent spliced precursors still bearing the 3′ extension (O’Connor and Peebles 1991). (C) The relative intensities of the hybridization signals in B were quantified by PhosphorImager analysis of the autoradiograph and plotted against the time of incubation at 36°C.