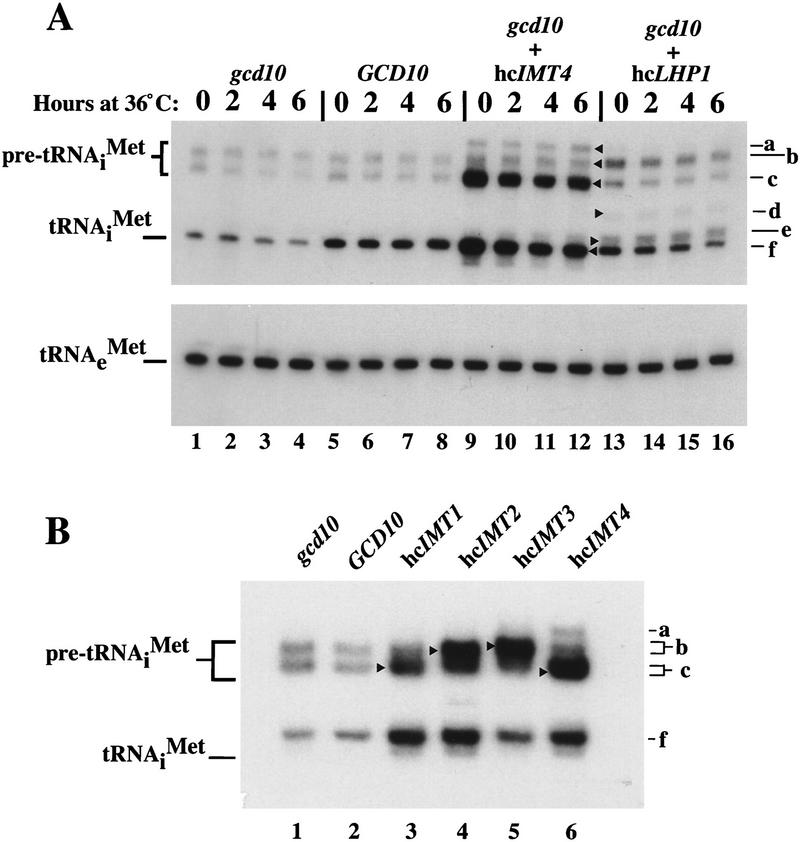
Figure 2.
High-copy suppressors of gcd10 mutations lead to increased amounts of initiator tRNAMet and identification of pre-tRNAiMet species. (A) Northern blot analysis of total RNA (5 μg) isolated from transformants of strains H2457 (gcd10-504) carrying empty vector YEp24 (gcd10 lanes), low-copy plasmid pMG107 bearing GCD10 (GCD10 lanes), high-copy plasmid pC44 bearing IMT4 (gcd10 + hcIMT4 lanes), or high-copy plasmid p2626 bearing LHP1 (gcd10 + hcLHP1 lanes), grown at 26°C or 36°C for the indicated times, as described in Fig. 1. The blot was probed for tRNAiMet and stripped and reprobed for tRNAeMet as described in Fig. 1. Indicated with arrowheads inside the blot and labeled to the right are the presumed positions of pre-tRNAiMet containing 5′ and 3′ extensions encoded by IMT4 and terminating downstream of the usual termination site (a), pre-tRNAiMet containing 5′ and 3′ extensions encoded by IMT2 and IMT3 (b), pre-tRNAiMet containing 5′ and 3′ extensions encoded by IMT1 and IMT4 (c), processing intermediate of IMT3 containing the 3′ extension (d), processing intermediate containing partial 3′ extension (e), and mature tRNAiMet (f). (B) Northern blot analysis of total RNA (10 μg) isolated from H2457 (gcd10-504) transformants containing empty vector YEp24 (gcd10), low-copy plasmid pMG107 bearing GCD10, or a high-copy plasmid bearing one of the four IMT genes, as indicated above the blot, grown at 26°C as described in Fig. 1. Initiator tRNAMet was detected by hybridization as described above and in Fig. 1. Indicated with arrowheads inside the blot and labeled to the right are the positions of pre-tRNAiMet containing 5′ and 3′ extensions (a–c) and mature tRNAiMet (f), as in A.