Figure 3.
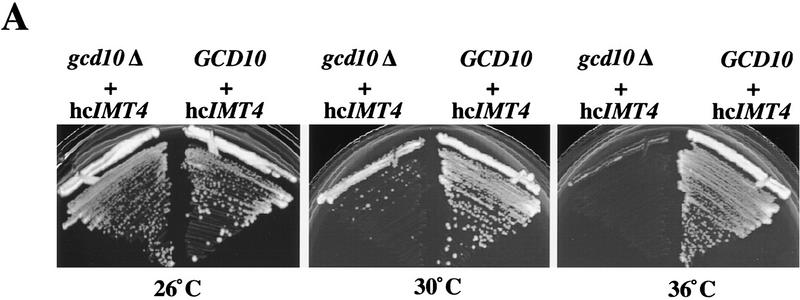
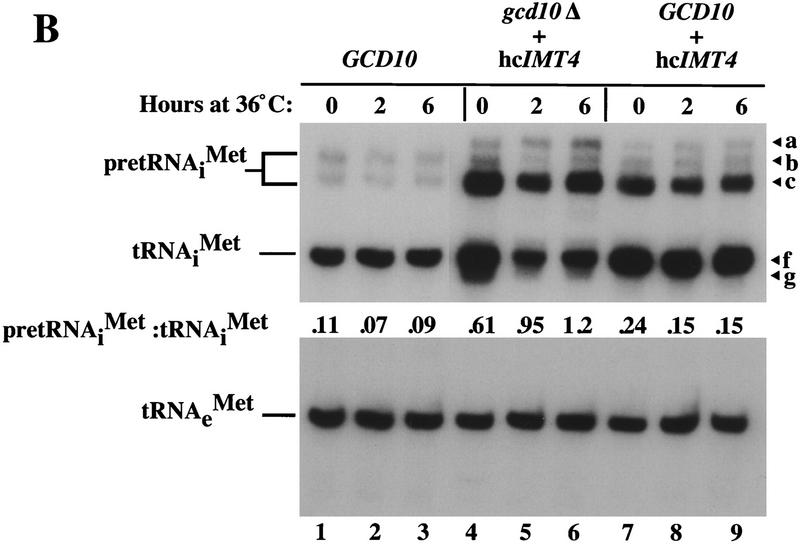
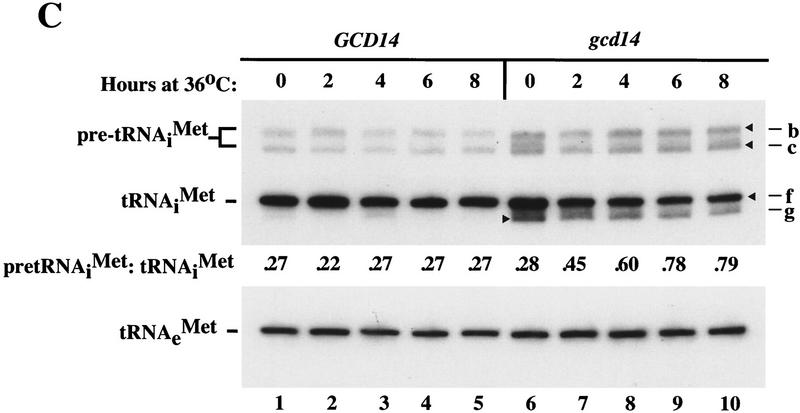
GCD10 is dispensable for cell viability in the presence of hcIMT4. (A) Growth of strain YJA146 (gcd10Δ + hcIMT4) and a transformant of its parental GCD10 strain BJ5464 bearing p1775 (GCD10 + hcIMT4) on YPD medium at 26°C for 3 days, and at 30°C or 36°C for 2 days. (B) Northern blot analysis of total RNA (7 μg) isolated from the same two strains described in A (gcd10Δ + hcIMT4 and GCD10 + hcIMT4 lanes) plus isogenic strain YJA143 containing the gcd10Δ chromosomal allele and single-copy GCD10 plasmid p2704 (GCD10). Strains were grown at 26°C or 36°C for 2 and 6 hr as described in Fig. 1. The membrane was probed for tRNAiMet and stripped and reprobed for tRNAeMet as described in Fig. 1. The different RNA species detected are indicated at left. The various forms of tRNAiMet species are labeled at right as in Fig. 2, with the addition of species g, which may be end-trimmed molecules lacking the CCA extension (see text). (C) Northern blot analysis of total RNA (5 μg) isolated from strain Hm296 (gcd14-2) containing wild-type GCD14 on single-copy plasmid pRC62 (GCD14 lanes) or empty vector YEp24 (gcd14 lanes), grown as described in Fig. 1. The membrane was probed for tRNAiMet and stripped and reprobed for tRNAeMet as described in Fig. 1. Indicated with arrowheads inside the blot and labeled to the right are the various tRNAiMet species described in Fig. 2 and above. The indicated ratios of pre-tRNAiMet to mature tRNAiMet were calculated from the relative intensities of hybridization signals quantitated by PhosphorImager analysis.