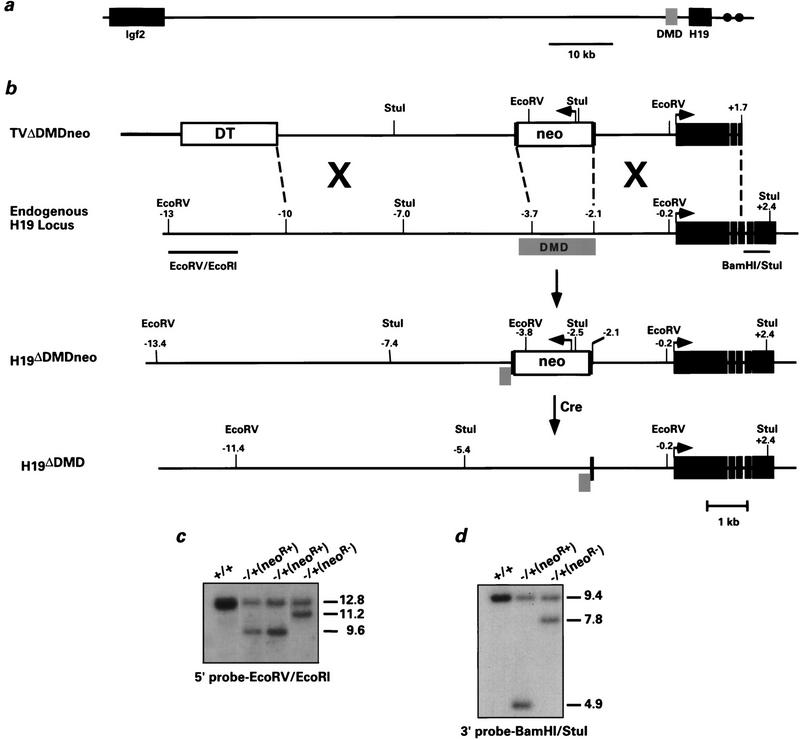
Figure 1.
Deletion of the H19 differentially methylated domain in ES cells. (a) The positions of Igf2 and H19 relative to the DMD on mouse chromosome 7 are indicated. The gray box corresponds to the 2-kb DMD, which is located −2 to −4 kb relative to the start of H19 transcription. (•) The endodermal enhancers located at +9 and +11 kb. (b) From top to bottom: the linearized targeting vector, the endogenous H19 locus, the targeted H19 locus (H19ΔDMDneo), and the targeted H19 locus after removal of PGK–neo using CRE–loxP recombination (H19ΔDMD). The linearized vector includes Bluescript II KS (thick line), the diphtheria toxin A gene (open box, DT), PGK–neo (open box, neo) flanked by loxP sites (black vertical bars), 5′ H19 sequence (thin black line) and H19 gene sequence extending into the third exon (solid boxes). Arrows indicate direction of transcriptional orientation of PGK–neo and H19. The positions of the restriction sites and the external probes (EcoRV/EcoRI and BamHI/StuI) used to analyze the targeted clones are indicated. (c,d) ES cell clones were screened by Southern blot analysis for the targeting event. Genomic DNA was digested with EcoRV and hybridized to the 5′ probe EcoRV/EcoRI (c) or with StuI and hybridized to the 3′ probe BamHI/StuI (d). The DNA samples shown include the parental ES cell DNA (+/+), targeted clones with the neor gene (neoR+), and a targeted clone in which the neor gene was excised (neoR−). Molecular sizes (in kb) are indicated to the right. Of the 85 G418-resistant clones analyzed, four were correctly targeted to the H19 locus.