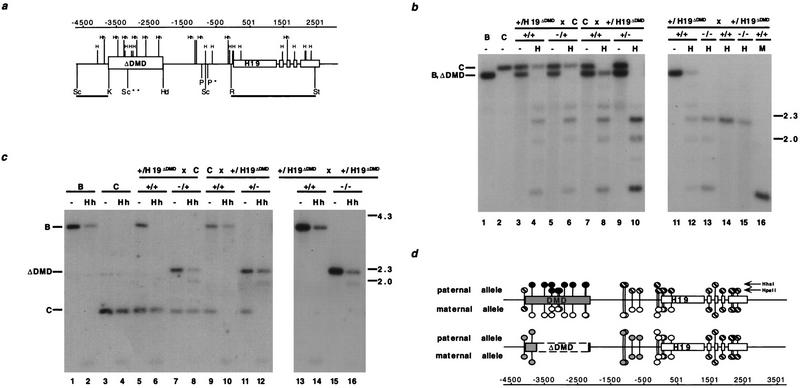
Figure 3.
Methylation analysis of H19 in heterozygous and homozygous DMD mutant mice. (a) The location of the HpaII (H) and HhaI (Hh) sites with respect to the deleted DMD sequence. The 5′ H19 DMD sequence, deleted between the KpnI (K) and HindIII (Hd) sites, and H19 are represented by boxed regions. (P) PvuII; (R) EcoRI; (St) StuI; (Sc) SacI. (*) The polymorphic PvuII site that is found in C57BL/6J and H19ΔDMD; (**) the polymorphic M. castaneus SacI site. The RSt probe used in b and the ScK probe used in c are indicated beneath the line. The position of the sites, relative to the start of transcription, are indicated (in bp) above the gene line. (b) The methylation status of HpaII sites in the H19 PvuII/StuI fragment is assessed with the RSt probe. DNA from C57BL/6J (B) mice, B6(CAST–H19) mice (C), progeny of a H19ΔDMD heterozygous female mated to a B6(CAST–H19) male (lanes 3–6), progeny from a B6(CAST–H19) female mated to a H19ΔDMD heterozygous male (lanes 7–10) and progeny of a H19ΔDMD heterozygous female mated to a H19ΔDMD heterozygous male (lanes 11–16) were analyzed. DNA was digested with PvuII and StuI and, in lanes indicated, HpaII (H) or MspI (M). The genotypes of the assayed DNA from the specific mating are indicated above the lanes. DNA from neonatal livers (lanes 1–13,16) and adult sperm (lanes 14,15) were assayed. The M. castaneus-specific PvuII/StuI fragment (C, 3.4 kb) and the C57BL/6 and H19ΔDMD-specific PvuII/StuI fragment (B, ΔDMD, 3.2 kb) are noted at left, with size markers in kb shown at right. (c) DNA from mice described in b was analyzed for HhaI methylation in the 5′ H19 SacI fragment using the ScK probe. DNA was digested with SacI and, in lanes indicated, HhaI (Hh). The genotypes of the DNA samples are indicated above the lanes. The C57BL/6J (3.8 kb), B6(CAST–H19) (1.5 kb) and the H19ΔDMD (2.2 kb)-specific SacI fragments are indicated at left. (d) Summary of the parental-specific methylation status of HpaII and HhaI sites on wild-type (top line) vs. H19ΔDMD (middle line) alleles. The taller and shorter lollipops represent the HhaI and the HpaII sites, respectively. (Solid circles) Fully methylated sites; (open circles) unmethylated sites; (striped circles) sites that are methylated on a subset of the alleles, as determined from previous studies (bisulfite sequence and Southern analyses) and data presented herewith; (shaded circles) sites that are partially methylated. The paternal-specific methylation status is presented above each allele; the maternal-specific methylation status is presented below each allele.