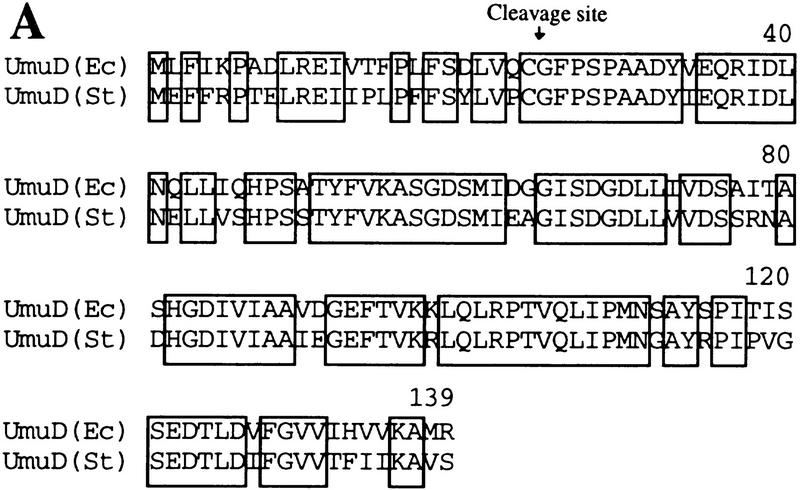
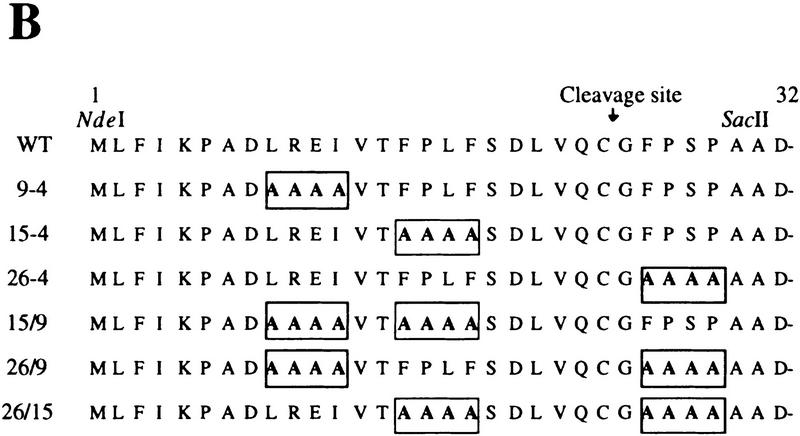
Figure 3.
Proteins used to identify the Lon recognition signal of UmuD. (A) E. coli and S. typhimurium UmuD proteins are both substrates of Lon (Gonzalez et al. 1998). Their primary amino acid sequences were aligned using the program Geneworks, and areas that are boxed indicate regions that are 100% conserved between the two proteins. Overall, the proteins are 73% identical, but most divergence occurs in the amino- and carboxy-terminal tails. (B) The amino terminus of the UmuD alanine-stretch mutants. Areas boxed indicate the multiple alanine substitutions made within the UmuD amino terminus for each mutant protein. The normal post-translational cleavage site is located between Cys-24 and Gly-25. The positions of the genetically engineered silent restriction sites for NdeI and SacII in the umuD gene are indicated at their approximate position in the corresponding UmuD protein.