Abstract
Plant VAPYRINs are required for the establishment of arbuscular mycorrhiza (AM) and root nodule symbiosis (RNS). In vapyrin mutants, the intracellular accommodation of AM fungi and rhizobia is blocked, and in the case of AM, the fungal endosymbiont cannot develop arbuscules which serve for nutrient exchange. VAPYRINs are plant-specific proteins that consists of a major sperm protein (MSP) domain and an ankyrin domain. Comparison of VAPYRINs of dicots, monocots and the moss Physcomitrella patens reveals a highly conserved domain structure. We focused our attention on the ankyrin domain, which closely resembles the D34 domain of human ankyrin R. Conserved residues within the petunia VAPYRIN cluster to a surface patch on the concave side of the crescent-shaped ankyrin domain, suggesting that this region may represent a conserved binding site involved in the formation of a protein complex with an essential function in intracellular accommodation of microbial endosymbionts.
Key words: VAPYRIN, arbuscular mycorrhiza, petunia, symbiosis, glomus, ankyrin, major sperm protein, VAP
Plants engage in mutualistic interactions such as root nodule symbiosis (RNS) with rhizobia and arbuscular mycorrhiza (AM) with Glomeromycotan fungi. These associations are referred to as endosymbioses because they involve transcellular passage through the epidermis and intracellular accommodation of the microbial partner within root cortical cells of the host.1,2 Infection by AM fungi and rhizobia is actively promoted by the plant and requires the establishment of infection structures namely the prepenetration apparatus (PPA) in AM and a preinfection thread in RNS, respectively.3–5 In both symbioses the intracellular microbial accommodation in epidermal and root cortical cells involves rebuilding of the cytoskeleton and of the entire membrane system.6–8 Recently, intracellular accommodation of rhizobia and AM fungi, and in particular morphogenesis of the AM fungal feeding structures, the arbuscules, was shown to depend on the novel VAPYRIN protein.9–11
VAPYRINs are plant-specific proteins consisting of two protein-protein interaction domains, an N-terminal major sperm protein (MSP) domain and a C-terminal ankyrin (ANK) domain. MSP of C. elegans forms a cytoskeletal network required for the motility of the ameboidal sperm.12 MSP domains also occur in VAP proteins that are involved in membrane fusion processes in various eukaryotes.13 The ANK domain, on the other hand, closely resembles animal ankyrins which serve to connect integral membrane proteins to elements of the spectrin cytoskeleton,14 thereby facilitating the assembly of functional membrane microdomains in diverse animal cells.15 Ankyrin repeats exhibit features of nano-springs, opening the possibility that ankyrin domains may be involved in mechanosensing.16 Based on these structural similarities, VAPYRIN may promote intracellular accommodation of endosymbionts by interacting with membranes and/or with the cytoskeleton. Indeed, VAPYRIN protein associates with small subcellular compartments in petunia and in Medicago truncatula.9,10
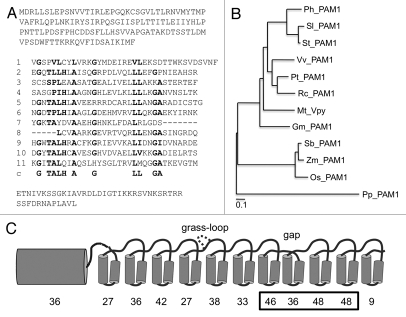
Ankyrin repeats typically consist of 33 amino acids, of which 30–40% are highly conserved across most taxa. These residues confer to the repeats their basic helix-turn-helix structure.17 Ankyrin domains often consist of arrays of several repeats that form a solenoid with a characteristic crescent shape.17 Besides the ankyrin-specific motiv-associated amino acids there is little conservation between the ankyrin domains of different proteins, or between the individual repeats of a given ankyrin domain,17 a feature that was also observed in petunia VAPYRIN (Fig. 1A).9 However, sequence comparison of VAPYRINs from eight dicots, three monocots and the moss Physcomitrella patens revealed a high degree of sequence conservation beyond the ankyrin-specific residues (Fig. 1B and Sup. Fig. S1). When the degree of conservation was determined for the individual ankyrin repeats among all the 12 species, it appeared that repeats 7, 9 and 10 exhibited particularly high conservation (Fig. 1C).
Figure 1.
Sequence analysis and phylogeny of VAPYRIN from diverse plants. (A) Predicted amino acid sequence of the petunia VAPYRIN protein PAM1. The 11 repeats of the ankyrin domain are aligned, and the ankyrin consensus sequence is shown below the eleventh ankyrin repeat (line c). Conserved residues that are characteristic for ankyrin repeats (Mosavi et al. 2004)17 are depicted in bold face. (B) Unrooted phylogenetic tree representing the VAPYRINs of eight dicot species (Petunia hybrida, Solanum lycopersicon, Solanum tuberosum, Vitis vinifera, Populus trichocarpa, Ricinus communis, Medicago truncatula and Glycine max) three monocot species (Sorghum bicolor, Zea mays and Oryza sativa), and the moss Physcomitrella patens. (C) Degree of conservation of the individual ankyrin repeats of VAPYRIN. Schematic representation of the MSP domain as N-terminal barrel-shaped structure, and of the individual ankyrin repeats as pairs of alpha-helices. An additional loop occurring only in monocots (grass-loop) is inserted above repeat 4, and the deletion between repeat 7 and 8 is indicated (gap). This latter feature is common to all VAPYRIN proteins. The percentage of amino acid residues that are identical in at least 11 of the 12 VAPYRINS is given below the MSP domain and the eleven ankyrin repeats. The box highlights repeats 7–10 which contribute to the predicted binding site (compare with Figs. 3 and 4).
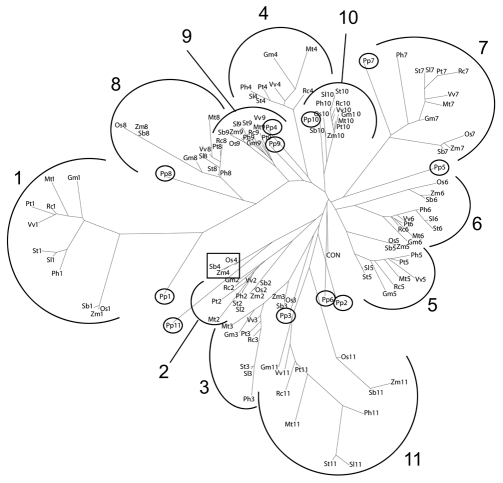
Sequence comparison of the eleven repeats of all the twelve plant species revealed that the individual repeats clustered according to their position in the domain, rather than according to their origin (plant species) (Fig. 2). This shows that the repeats each are well conserved across species, but show little similarity among each other within a given VAPYRIN protein. The higher conservation of repeats 9 and 10 was reflected by the compact appearance of the respective branches, in which the monocot and moss sequences were nested closely with the dicot sequences, compared to other repeats, where the branches appeared fragmented between monocots and dicots, and where the P. patens sequence fell out of the branch as in the case of repeats 4–6 (Fig. 2). Taken together, this points to an old evolutionary origin of the entire ankyrin domain in lower land plants, with no subsequent rearrangement of ankyrin repeats.
Figure 2.
Phylogenetic analysis of the individual ankyrin repeats of VAPYRIN. Phylogenetic representation of an alignment of all the 11 repeats of the 12 VAPYRINs compared in Figure 1B and C. The repeats cluster according to their position within the domain, rather than to their origin (plant species). Numbers indicate the position of the repeats within the domain (compare with Fig. 1C). P. patens repeats are highlighted (small circles) for clarity. The monocot repeat 4 sequences (boxed) are remote from the remaining repeat 4 sequences because of the grass loop (compare with Fig. 1C).
Ankyrin domains function as protein-protein interaction domains,17 in which the residues on the surface are involved in the binding of their protein partners.14 The fact that repeats 9 and 10 exhibited particularly high levels of conservation across species from moss to angiosperms indicated that this region may contain functionally important residues. Within repeat 10, sixteen amino acid positions were identical in >90% of the analyzed species (Fig. 3A and grey bars). Nine of those represent residues that are characteristic for ankyrin repeats (red letters) and determine their typical 3D shape.17 These residues are considered ankyrin-specific, and are unlikely to be involved in a VAPYRIN-specific function. The remaining seven highly conserved residues in repeat 10, however, are VAPYRIN-specific, since they have been under positive selection, without being essential for the basic structure of the ankyrin repeat. Ankyrin-specific and VAPYRIN-specific residues where identified throughout the entire ankyrin domain (Sup. Fig. 1), and subsequently mapped on a 3-dimensional model of petunia VAPYRIN to reveal their position in the protein (Fig. 3B–G). The ankyrin-specific residues were found to be localized primarily to the interior of the ankyrin domain, with the characteristic glycines (brown) marking the turns between helices and loops (Fig. 3B, D and F, compare with A). In contrast, the VAPYRIN-specific residues were localized primarily on the surface of the ankyrin domain (Fig. 3C, E and G). A prominent clustering of VAPYRIN-specific residues was identified on the concave side of the crescent-shaped ankyrin domain comprising repeats 7–10 close to the gap (Figs. 3G and 4). This highly conserved VAPYRIN-specific region contains several negatively and positively charged residues (D, E and K, R, respectively) and aromatic residues (W, Y, F), which may together form a conserved binding site for an interacting protein.
Figure 3.
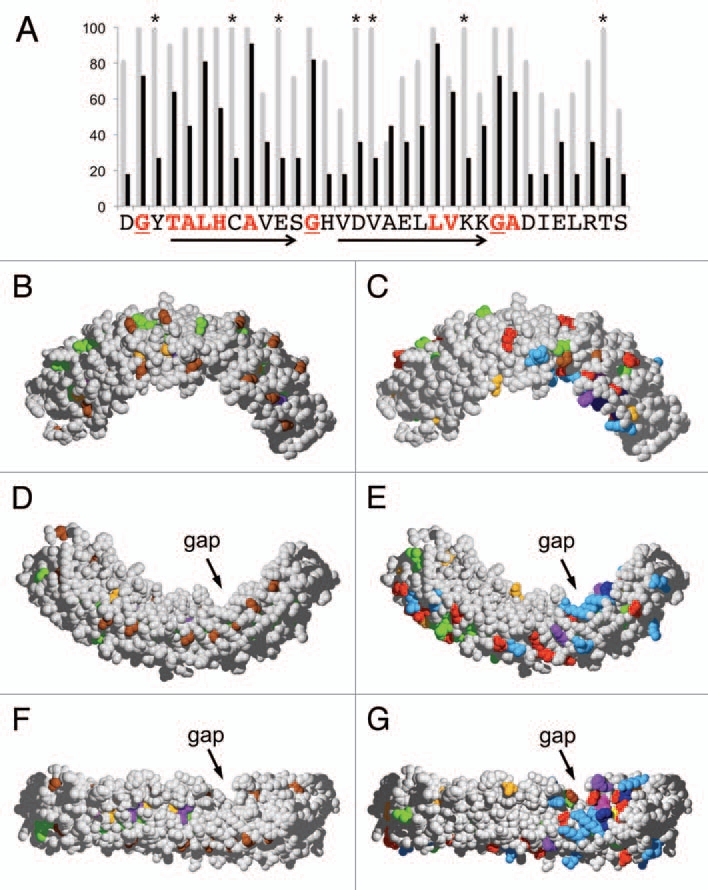
3D-Mapping of conserved positions within the ankyrin domain of VAPYRIN. (A) Conserved amino acid residues were evaluated for ankyrin repeat 10 of petunia VAPYRIN as an example. The degree of conservation between the 12 VAPYRINs analyzed in Figures 1B and 2 is depicted with grey bars. Average conservation between all the 132 ankyrin repeats of the 12 VAPYRIN sequences is shown with black bars. Residues that are conserved in all 132 repeats (red letters) define the ankyrin consensus sequence, which confers to the repeats their characteristic basic structure.17 Residues that are >90% conserved but are not part of the basic ankyrin sequence (highlighted with asterisks) are VAPYRIN-specific and may therefore have been conserved because of their specific function in VAPYRIN. Arrows indicate the characteristic antiparallel helices, the turns are marked by conserved glycine residues (underlined; compare with B, D and F). (B–G) 3D-models of the petunia VAPYRIN PAM1. Conserved amino acid residues were color-coded according to their physico-chemical properties (http://life.nthu.edu.tw/∼fmhsu/rasframe/SHAPELY.HTM) with minor modification (see below). In (B, D and F) the ankyrin-specific residues are highlighted (corresponding to the bold letters in Fig. 1A). In (C, E and G), the VAPYRIN-specific residues are highlighted. Note the patch of high conservation on the concave side of the crescent-shaped ankyrin domain between repeats 7–10 next to the gap. (B–E) represent respective side views of the ankyrin domain, (F and G) exhibit the concave inner side of the domain. Color code: Bright red: aspartic acid (D), glutamic acid (E); Yellow: cysteine (C); Blue: lysine (K), arginine (R); Orange: serine (S), threonine (T); Dark blue: phenylalanine (F), tyrosin (Y); Brown: glycine (G); Green: leucin (L), valine (V), isoleucin (I), alanine (A); Lilac: tryptophane (W); Purple: histidine (H); Pink: proline (P).
Figure 4.
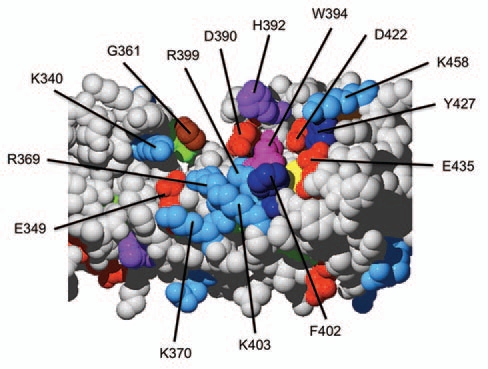
The highly conserved surface area in domain 8–10 of the ankyrin domain of petunia VAPYRIN. Close-up of the highly conserved region of petunia PAM1 as shown in Figure 3G. Amino acids were color-coded as in Figure 3 and their position in the amino acid sequence is indicated (compare with Sup. Fig. 1).
In this context, it is interesting to note that human ankyrin R also contains a binding surface on the concave side of the D34 domain for the interaction with the CBD3 protein.14 Consistent with an essential function of the C-terminal third of the ankyrin domain, mutations that abolish this relatively short portion of VAPYRIN, have a strong phenotype, indicating that they may represent null alleles.9 Based on this collective evidence, we hypothesize that repeats 7–10 are involved in the formation of a protein complex that is essential for intracellular accommodation of rhizobia and AM fungi. Biochemical and genetic studies are now required to identify the binding partners of VAPYRINs, and to elucidate their role in plant endosymbioses.
Supplementary Material
References
- 1.Oldroyd GED, Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. Ann Rev Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- 2.Parniske M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat Rev Microbiol. 2008;6:763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- 3.Genre A, Chabaud M, Faccio A, Barker DG, Bonfante P. Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell. 2008;20:1407–1420. doi: 10.1105/tpc.108.059014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG. Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell. 2005;17:3489–3499. doi: 10.1105/tpc.105.035410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fournier J, Timmers ACJ, Sieberer BJ, Jauneau A, Chabaud M, Barker DG. Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol. 2008;148:1985–1995. doi: 10.1104/pp.108.125674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genre A, Bonfante P. Actin versus tubulin configuration in arbuscule-containing cells from mycorrhizal tobacco roots. New Phytol. 1998;140:745–752. doi: 10.1046/j.1469-8137.1998.00314.x. [DOI] [PubMed] [Google Scholar]
- 7.Genre A, Bonfante P. Epidermal cells of a symbiosis-defective mutant of Lotus japonicus show altered cytoskeleton organisation in the presence of a mycorrhizal fungus. Protoplasma. 2002;219:43–50. doi: 10.1007/s007090200004. [DOI] [PubMed] [Google Scholar]
- 8.Blancaflor EB, Zhao LM, Harrison MJ. Microtubule organization in root cells of Medicago truncatula during development of an arbuscular mycorrhizal symbiosis with Glomus versiforme. Protoplasma. 2001;217:154–165. doi: 10.1007/BF01283396. [DOI] [PubMed] [Google Scholar]
- 9.Feddermann N, Duvvuru Muni RR, Zeier Z, Stuurman J, Ercolin F, Schorderet M, et al. The PAM1 gene of petunia, required for intracellular accommodation and morphogenesis of arbuscular mycorrhizal fungi, encodes a homologue of VAPYRIN. Plant J. 2010;64:470–481. doi: 10.1111/j.1365-313X.2010.04341.x. [DOI] [PubMed] [Google Scholar]
- 10.Pumplin N, Mondo SJ, Topp S, Starker CG, Gantt JS, Harrison MJ. Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J. 2010;61:482–494. doi: 10.1111/j.1365-313X.2009.04072.x. [DOI] [PubMed] [Google Scholar]
- 11.Murray JD, Duvvuru Muni R, Torres-Jerez I, Tang Y, Allen S, Andriankaja M, et al. Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 2011;65:244–252. doi: 10.1111/j.1365-313X.2010.04415.x. [DOI] [PubMed] [Google Scholar]
- 12.Italiano JE, Roberts TM, Stewart M, Fontana CA. Reconstitution in vitro of the motile apparatus from the amoeboid sperm of Ascaris shows that filament assembly and bundling move membranes. Cell. 1996;84:105–114. doi: 10.1016/s0092-8674(00)80997-6. [DOI] [PubMed] [Google Scholar]
- 13.Lev S, Ben Halevy D, Peretti D, Dahan N. The VAP protein family: From cellular functions to motor neuron disease. Trends Cell Biol. 2008;18:282–290. doi: 10.1016/j.tcb.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 14.Michaely P, Tomchick DR, Machius M, Anderson RGW. Crystal structure of a 12 ANK repeat stack from human ankyrinR. EMBO J. 2002;21:6387–6396. doi: 10.1093/emboj/cdf651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett V, Healy J. Membrane domains based on ankyrin and spectrin associated with cell-cell interactions. Cold Spring Harb Perspect Biol. 2009 doi: 10.1101/cshperspect.a003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee G, Abdi K, Jiang Y, Michaely P, Bennett V, Marszalek PE. Nanospring behavior of ankyrin repeats. Nature. 2006;440:246–249. doi: 10.1038/nature04437. [DOI] [PubMed] [Google Scholar]
- 17.Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.