Figure 3.
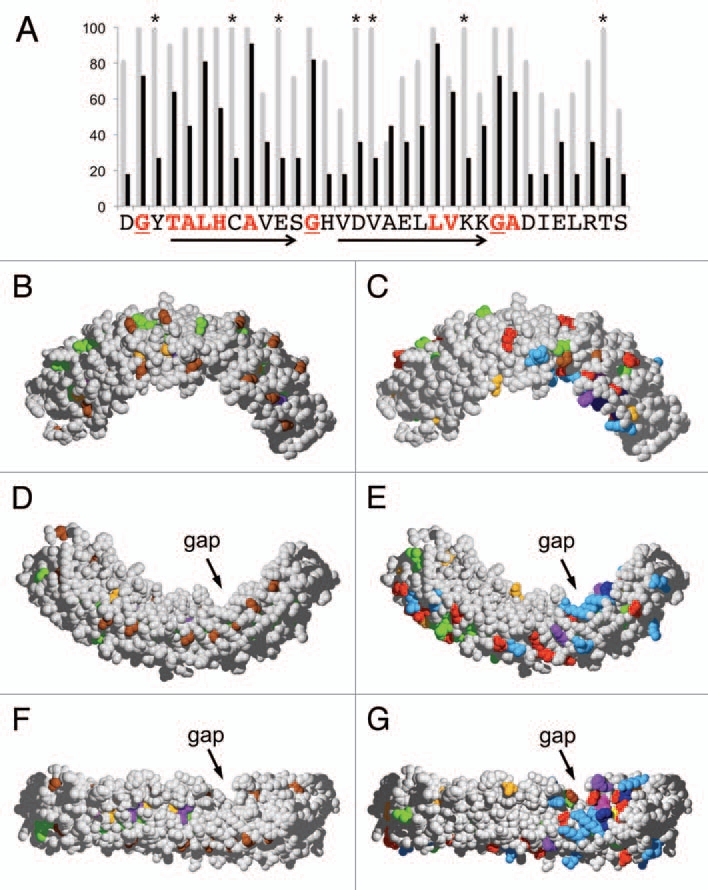
3D-Mapping of conserved positions within the ankyrin domain of VAPYRIN. (A) Conserved amino acid residues were evaluated for ankyrin repeat 10 of petunia VAPYRIN as an example. The degree of conservation between the 12 VAPYRINs analyzed in Figures 1B and 2 is depicted with grey bars. Average conservation between all the 132 ankyrin repeats of the 12 VAPYRIN sequences is shown with black bars. Residues that are conserved in all 132 repeats (red letters) define the ankyrin consensus sequence, which confers to the repeats their characteristic basic structure.17 Residues that are >90% conserved but are not part of the basic ankyrin sequence (highlighted with asterisks) are VAPYRIN-specific and may therefore have been conserved because of their specific function in VAPYRIN. Arrows indicate the characteristic antiparallel helices, the turns are marked by conserved glycine residues (underlined; compare with B, D and F). (B–G) 3D-models of the petunia VAPYRIN PAM1. Conserved amino acid residues were color-coded according to their physico-chemical properties (http://life.nthu.edu.tw/∼fmhsu/rasframe/SHAPELY.HTM) with minor modification (see below). In (B, D and F) the ankyrin-specific residues are highlighted (corresponding to the bold letters in Fig. 1A). In (C, E and G), the VAPYRIN-specific residues are highlighted. Note the patch of high conservation on the concave side of the crescent-shaped ankyrin domain between repeats 7–10 next to the gap. (B–E) represent respective side views of the ankyrin domain, (F and G) exhibit the concave inner side of the domain. Color code: Bright red: aspartic acid (D), glutamic acid (E); Yellow: cysteine (C); Blue: lysine (K), arginine (R); Orange: serine (S), threonine (T); Dark blue: phenylalanine (F), tyrosin (Y); Brown: glycine (G); Green: leucin (L), valine (V), isoleucin (I), alanine (A); Lilac: tryptophane (W); Purple: histidine (H); Pink: proline (P).
