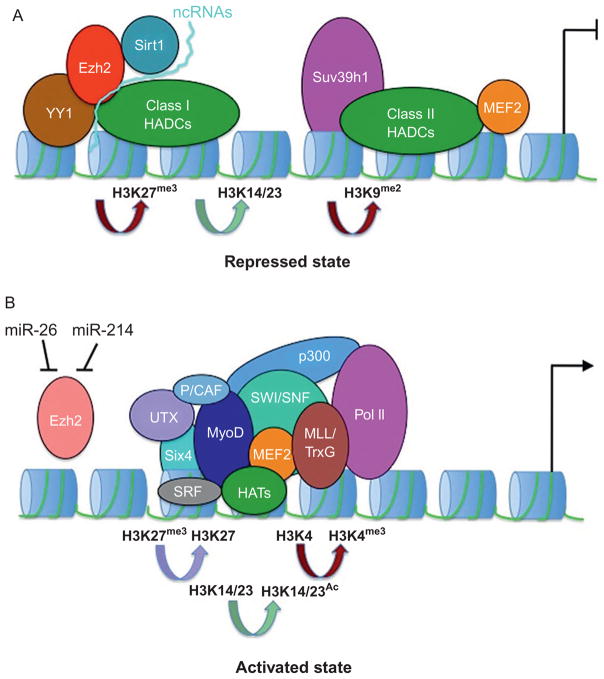
Figure 3.1.
(A) Repressive histone marks are introduced by protein complexes composed by class I, II, and III (SIRT1) deacetylases and histone methyltransferases (Suv39h1 and Polycomb Ezh2). Ezh2 may be recruited by the Pleiohomeotic (PHO)-related YY1 protein or noncoding RNAs (ncRNAs). Lysines 14–23 of histone H3 are deacetylated and lysine 27 (Ezh2) and lysine 9 (Suv39h1) are methylated. (B) Repressors are replaced by a new set of protein complexes at transcribed genes. Via interaction with the transcriptional activators MyoD, MEF2, SRF, and Six4, acetyltransferases (HATs, p300, P/CAF) and nucleosome remodeling machines (SWI/SNF) promote lysine acetylation (H3K14-23AC) and nucleosome remodeling, respectively. Demethylation of lysine 27 is achieved by transcriptional downregulation and microRNA-mediated (miR-26 and miR-214) repression of Ezh2. In addition, the lysine 27-specific UTX actively removes methyl groups from lysine 27. The Trithorax-related mixed lineage leukemia (MLL) proteins promote methylation of histone H3 lysine 4, a mark associated with polymerase II (PolII) recruitment and gene activation.