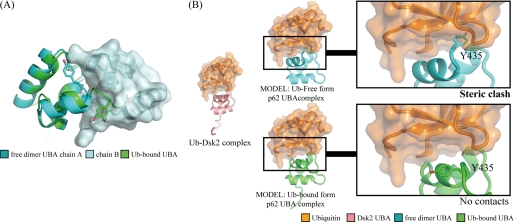
FIGURE 5.
Dimerization and ubiquitin binding of the p62 UBA domain. A, overlay of the free-form dimer crystal structure and the ubiquitin (Ub)-bound NMR structure of p62 UBA. The extended helix-3 that is formed upon binding to ubiquitin crashes to the other p62 UBA domain when p62 UBA is in a dimer configuration. B, model structures of the free-form crystal structure (cyan) and the ubiquitin-bound NMR structure (green) of p62 UBA complexed with ubiquitin (orange). The models were built based on the complex structure between Dsk2 UBA and ubiquitin (Protein Data Bank code 1WR1, pink and orange). Backbone atoms of the UBA domain from the complex were superimposed onto p62 UBA (cyan and green). The Dsk2 UBA domain from the original complexes was then removed. The C-terminal tail region, including the Tyr435 side chain, which has been shown to undergo a critical homomeric interaction for dimerization in this study, crashes with ubiquitin in the complex-form model (top), whereas no crashing is observed in the model generated from ubiquitin-bound form p62 UBA structure. These models suggest that the dimerization and ubiquitin binding are mutually exclusive.