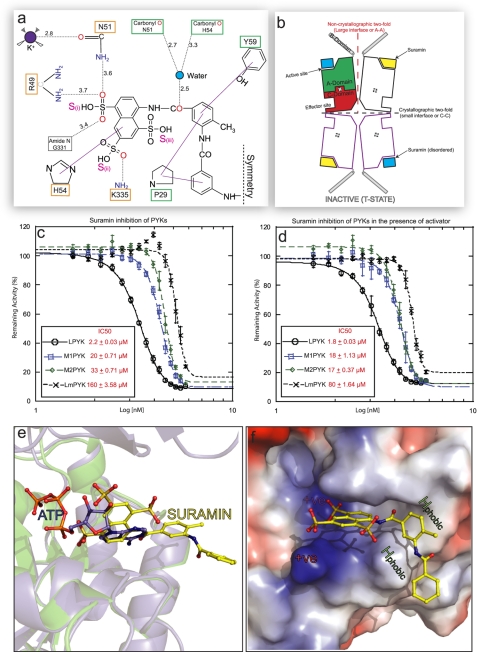
FIGURE 3.
Suramin inhibition of PYK is enhanced in the presence of the activator molecule. a, a schematic drawing showing the interatomic distances (given in Å, dotted black lines) for the interactions shown in Fig. 2b). The solid purple lines indicate stacking interactions. Residues involved in ATP binding (orange box) and not involved (green) have been indicated. Only one symmetrical half of the suramin molecule is shown (as indicated by the dashed symmetry line). b, schematic representation of the inactive LmPYK-suramin structure. Suramin is unambiguously (yellow lozenges) bound to one of the chains in the asymmetric unit (two chains outlined in black) but is disordered (blue square) in the adjacent chain, separated by the large (A-A) interface. A crystallographic two-fold axis running along the small interface generates two more chains (purple outlines), forming the biologically relevant tetramer structure. See Ref. 20 for schematics of the enzyme in its other conformations. c and d, concentration-response curves observed for the titration of suramin against PYK activity in the absence (c) and presence (d) of activator (F16BP for human PYKs, F26BP for LmPYK), using a luciferase-based assay. The values are expressed ± S.D. Suramin showed no inhibition in the control assay of luciferase activity alone. All assays were performed in triplicate. e, the binding of suramin prevents ADP/ATP binding in trypanosomatid PYKs. The R-state LmPYK-ATP/oxalate/F26BP monomer (colored blue, PDB ID = 3HQP) was superposed onto the inactive LmPYK-suramin structure (colored green). The A- and C-domains (residues 18–86 and 188–480) of both structures were superposed (r.m.s. fit of the C-α atoms of domains A and C is 0.50 Å). The superpositions of the ATP and suramin molecules clash, indicating a clear mechanism of competitive inhibition. f, the electrostatic surface of the suramin binding site, showing areas of positive charge (blue), which interact with the negatively charged sulfono groups of suramin. Two hydrophobic groups (Pro-29 and Tyr-59) provide further stability, helping to hold the molecule in the ATP/ADP binding site.