Abstract
Several factors regulate nerve growth factor (NGF), which is formed from pro-NGF by intracellular and extracellular enzymatic cleavage. The close proximity between mast cells expressing the protease tryptase and NGF-producing smooth muscle-like peritubular cells in the testes of infertile patients led us to examine whether tryptase is among those factors. Human peritubular cells express functional tryptase receptors (PAR-2). Recombinant enzymatically active β-tryptase increased NGF levels in the culture medium of primary human peritubular cells, but the peptide agonist for PAR-2 (SLIGKV) did not. Neither tryptase nor the peptide increased NGF mRNA levels. To test whether the increase in NGF is due to enzymatic activity of tryptase acting on pro-NGF, supernatants of peritubular cells and synthetic pro-NGF were treated with tryptase. Results of Western blot studies indicate enzymatic cleavage of pro-NGF by active tryptase. Heat-inactivated tryptase or SLIGKV was not effective. Mass spectrometry analysis of in vitro cleavage products from recombinant tryptase and synthetic pro-NGF revealed multiple cleavage sites within the pro-NGF sequence. The results also indicate the generation of mature NGF and smaller NGF fragments as a result of tryptase action. Thus, tryptase-secreting mast cells in the vicinity of pro-NGF/NGF-secreting cells in any human tissue are likely able to alter the ratios of pro-NGF/NGF. As NGF and pro-NGF have different affinities for their receptors, this indicates a novel way by which mast cells, via tryptase, can modify the microenvironment in human tissues with regard to neurotrophin actions.
Keywords: Mast Cell, Neurobiology, Neurotrophic Factor, Protease, Smooth Muscle, Human, Pro-NGF, Testis
Introduction
Expression and secretion of the prototype neurotrophin nerve growth factor (NGF) can be regulated by different factors. Forskolin, interleukin-1 (IL-1), and platelet-derived growth factor (PDGF) are able to increase the level of NGF mRNA (1), catecholamine, histamine, lipopolysaccharide (LPS), and tumor necrosis factor (TNF)-α for example can regulate synthesis and/or secretion of NGF (2–4).
Neurotrophins, including NGF, are produced as precursor molecules. The pro regions of neurotrophins appear to be important for correct folding and direct neurotrophins to the regulated secretory pathway (5, 6). Proneurotrophins can be cleaved intracellularly by furin or proconvertases (7, 8), whereas after secretion, extracellular plasmin or matrix metalloproteinase type 7 is responsible for cleaving pro-NGF. NGF finally is degraded by plasmin-activated matrix metalloproteinase 9 (9).
NGF binds two classes of receptors, the common low affinity p75NTR and the more selective high affinity tropomyosin-related kinase (Trk A) receptor. Activation of these by NGF has been linked to life or death of target cells (10–12). Recently, pro-NGF has also been implicated in the regulation of cell fate. Rather than being an inactive precursor, pro-NGF was found to bind to sortilin, which interacts and binds p75NTR and thus links this molecule to cell death (8, 13, 14). It appears that the ratio of pro-NGF to NGF and the presence of receptors (TrkA/p75NTR and sortilin) determine the balance between life and apoptosis (15) at least in typical neuronal targets.
It is clear by now that NGF is expressed not only by neuronal tissues (e.g. sympathetic neurons (16) and dorsal root ganglia (17)) but by many different tissues outside the nervous system. These include cells of the ovary (18–20), bone marrow (21), hair follicles (22), liver (23), urinary bladder (24), retina (25), prostate cells (26), oral mucosal keratinocytes (27), and spermatids (28). NGF is also produced and released by cells of the immune system, namely mast cells, which also respond to NGF (29, 30).
Results of our previous studies showed that the smooth muscle-like peritubular cells, which form the cellular components of the wall of the seminiferous tubule in man, are among the non-neuronal cell types able to produce NGF (31) as well as glial cell line-derived neurotrophic factor (32). The secretion of both factors occurs constitutively but may be further regulated by local factors, which can be derived from immune cells.
At least two types of immune cells accumulate in the peritubular wall of men with fertility problems, namely mast cells (see Fig. 1) and macrophages, and both are a source of TNF-α (33–35). When this cytokine was added to isolated human testicular peritubular cells (HTPCs),3 NGF mRNA and NGF protein production and accumulation in the medium increased (31). However, TNF-α failed to affect glial cell line-derived neurotrophic factor produced by HTPCs and equivalent cells isolated from the testes of infertile men (HTPC-Fs; (32)).
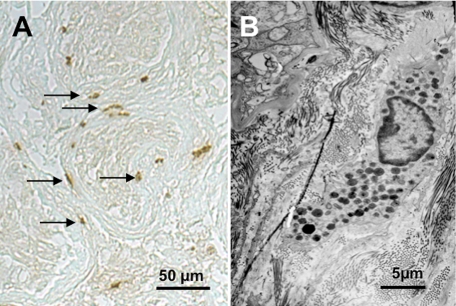
FIGURE 1.
Mast cells are abundant in infertile men. Mast cells are encountered in human testicular biopsies from men with impaired spermatogenesis (for details, see Ref. 33). They are identified by immunohistochemistry and contain immunoreactive tryptase (A). These cells accumulate in the peritubular compartment (arrows in A) and show the typical features of mast cells, including electron-dense vesicles at the ultrastructural level (B).
Likewise, tryptase, the major mast cell product found in all human mast cells, including testicular mast cells (see Fig. 1 and Ref. 36)), did not alter glial cell line-derived neurotrophic factor levels (32), although functional tryptase receptors (protease-activated receptor-2 (PAR-2)) are present (37). Whether tryptase may be able to affect NGF production of testicular peritubular cells or any other cell types is not known. Tryptase is a serine protease with multiple known functions, including the activation of the cell surface receptor PAR-2, a G-protein-coupled receptor expressed by many cells, including human testicular peritubular cells (32, 38, 39). Proteolytic cleavage is necessary for PAR-2 activation and signaling. However, human mast cell tryptase is also known to cleave a number of extracellular proteins, thereby catalyzing the activation of complement C3, converting prostromelysin to stromelysin (MMP3), whereas cleavage of fibrinogen results in a loss of clotting potential (40, 41). Tryptase furthermore degrades fibronectin, calcitonin gene-related peptide, vasoactive intestinal peptide, kininogen (42–48), and IgE (49). Mast cells via the enzymatic actions of tryptase therefore are able to regulate functions of PAR-2-expressing target cells and in addition are able to control different aspects of the local microenvironment. Based on the in vivo situation in the peritubular wall of male infertility patients in which tryptase-positive mast cells are in close contact with peritubular cells, we examined the ability of tryptase to regulate pro-NGF/NGF secretion and processing using primary HTPCs and HTPC-Fs.
EXPERIMENTAL PROCEDURES
Isolation of HTPC/-Fs, Cell Culture, and Human Testicular Samples
Isolation of HTPC/-Fs was performed as described (31, 32, 38). All participants granted written informed consent. The local ethics committee approved the study and the use of the cells. Patients displayed either normal spermatogenesis or impaired spermatogenesis and testicular fibrosis based on histological analyses. Cells were cultured in DMEM + 10% fetal calf serum (FCS; both from PAA GmbH, Cölbe, Germany). Treatment with 10 and 100 μm SLIGKV (NeoMPS, Strasbourg, France) or 10, 100, and 1000 ng/ml human recombinant skin β-tryptase (Promega, Mannheim, Germany) was performed as detailed below. For all experiments, freshly isolated or cryopreserved cells from passages 5–12 were used.
Immunocytochemistry
Immunofluorescence methods were performed as described (38, 50) with the following antisera: pro-NGF polyclonal antiserum (Alomone, Jerusalem, Israel; 1:50; rabbit) and NGF polyclonal antiserum (Chemicon Inc., Temecula, CA; 1:100; rabbit). Controls consisted of incubation with non-immune normal serum (rabbit; 1:5000) instead of specific antibodies or omission of the primary antibody.
Isolation of RNA and RT-PCR
Cells from at least four patients per experiment were grown to subconfluence, washed twice with PBS, and suspended in RLT buffer (Qiagen GmbH, Hilden, Germany) containing 1% β-mercaptoethanol (according to the manufacturer's protocol). Isolation of RNA (Qiagen RNeasy minikit) was followed by reverse transcription using random 15-mer primers. Four hundred nanograms of total RNA were used for further RT-PCR experiments (32, 34, 35). PCR products were visualized by ethidium bromide staining in agarose gels. Negative controls (e.g. omitting the respective input cDNA) were performed. All primers spanned at least one intron. The following primers were used: NGF, forward sense primer 5′-AGG GAG CAG CTT TCT ATC CTG-3′ and reverse antisense primer 5′-GGC AGT GTC AAG GGA ATG C-3′ yielding a 185-bp fragment; and cyclophilin, forward sense primer 5′-CTC CTT TGA GCT GTT TGC AG-3′ and reverse antisense primer 5′-CAC CAC ATG CTT GCC ATC C-3′ yielding a 325-bp fragment. Identities of PCR products were verified by sequencing (34, 35). For semiquantitative studies, intensities of bands were evaluated as described (51) using Image J (National Institutes of Health, Bethesda, MD; version 1.37) and normalized to those of cyclophilin.
Real Time Quantitative PCR (qRT-PCR)
Expression levels of NGF (forward sense primer 5′-AGC TTT CTA TCC TGG CCA CA-3′ and reverse antisense primer 5′-CAG TGT CAA GGG AAT GCT GA-3′) were quantified using qRT-PCR with RNA from four different patients. The primer pairs were intron-spanning to exclude co-amplification of genomic DNA. The cDNA was diluted 1:50, and duplicates were used to quantify the expression of NGF in each cDNA sample using a DyNAmo HS SYBR Green qPCR kit (Finnzymes, Espoo, Finland). Expression levels of the respective gene were normalized to the level of the ribosomal housekeeping gene L19 (RPL19) (52, 53).
Western Blotting
HTPC/-Fs were seeded on 60-mm dishes (Nunc GmbH & Co. KG, Wiesbaden, Germany) and incubated in 2 ml of DMEM without FCS with/without tryptase (1000 ng/ml), heat-inactivated tryptase (1000 ng/ml; 15 min at 95 °C), or the PAR-2 agonist peptide SLIGKV (100 μm) for 24 h. For concentration and purification of the supernatants, centrifugal filters (Amicon Ultra-4 3000, Millipore, Cork, Ireland) were used. For in vitro experiments, synthetic human pro-NGF (ProSpec-Tany TechnoGene Ltd., Israel; Lot Number 808PRONGF01) was treated with/without recombinant human tryptase (1000 ng/ml), heat-inactivated tryptase (1000 ng/ml; 15 min at 95 °C), or SLIGKV (100 μm) for 24 h at 37 °C. Immunoblotting was performed as described (34) using the same specific antisera as for immunocytochemistry (anti-human pro-NGF antiserum at 1:400 and anti-human NGF antiserum at 1:500). All Western blots were repeated with cells or supernatants from 6–12 different patients. For preadsorption of pro-NGF antiserum, equal amounts of antiserum and peptide (provided by Alomone) were incubated at room temperature for 2 h (n = 2). After centrifugation, the supernatant was used for the incubation. Intensities of bands were evaluated as described (51) using Image J.
Cell Viability Assay
Cell viability was estimated by measuring ATP content, which correlates with cell number and/or viability (54, 55). For the ATP assay, cells were cultured on 24-well plates (Nunc GmbH & Co. KG) and treated with/without 1000 ng/ml tryptase, 1000 ng/ml heat-inactivated tryptase (15 min at 95 °C), or 100 μm SLIGKV for 24 h. For the reaction, 100 μl of media plus 100 μl of CellTiter-Glo® reagent (Promega, Mannheim, Germany) were added. In the dark, contents of wells were mixed on a plate shaker at 500 rpm for 2 min and then incubated for 10 min. The luminescence of each sample was measured on a white 96-well plate (Nunc GmbH & Co. KG) in a plate-reading luminometer (Fluostar Optima, BMG Labtech, Offenburg, Germany). All measurements were performed in quadruples with cells from seven different patients.
NGF ELISA
Cells were stimulated with tryptase (10, 100, and 1000 ng/ml) or the PAR-2 agonist peptide SLIGKV (10 and 100 μm) in DMEM without serum, and supernatants were collected after 24 h. These concentrations were chosen because 10 and 1000 ng/ml tryptase and 10 and 100 μm SLIGKV robustly induce increases in intracellular calcium levels indicative of activation of PAR-2 (not shown). All samples for this assay were prepared under serum-free conditions to prevent interference of serum contents with NGF detection as recommended by the manufacturer. All samples were stored at −20 °C until use. Measurements of secreted NGF levels were performed using the NGF Emax ImmunoAssay System (Promega, Madison, WI) according to the manufacturer's protocol. Briefly, Nunc MaxiSorp 96-well plates (Nunc GmbH & Co. KG) were coated overnight with polyclonal NGF antiserum. The following day samples were added and captured by a monoclonal antibody against NGF followed by signal detection. Absorbance was measured at 450 nm using a Fluostar photometer (Fluostar Optima, BMG Labtech). All measurements were performed at least in duplicate with cells from 10 different patients and were normalized to cellular protein (31, 34).
LC-MS/MS Analyses
As for Western blotting experiments, 4 μl of sample containing 200 ng of synthetic human pro-NGF (treated for 24 h at 37 °C with 1000 ng/ml recombinant human tryptase) was reduced by addition of 0.4 μl of 45 mm DTT and incubation at 55 °C for 30 min. Alkylation was performed by addition of 0.4 μl of 0.1 m iodoacetamide and incubation at RT for 30 min. Samples were diluted in 0.1% formic acid to a final volume of 40 μl and subjected to LC-MS/MS analysis. Chromatographic separation of peptides was performed using a nano-HPLC system (Ettan MDLC, GE Healthcare Europe GmbH, Freiburg, Germany). Samples were loaded on a reversed phase trap column at a flow rate of 10 μl/min (loading buffer, 0.1% formic acid; trap column, C18 PepMap 100, 5-μm bead size, 300-μm inner diameter, 5-mm length (LC Packings, Sunnyvale, CA)) and subsequently separated on an analytical column (Reprosil-Pur C18 AQ, 3 μm, 150 mm × 75 μm (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany)). Solvent A consisted of 0.1% formic acid, and solvent B was composed of 84% acetonitrile in 0.1% formic acid. Separation was performed at a flow rate of 280 nl/min and an 80-min gradient from 0% B to 30% B followed by a 30-min gradient from 30% B to 60% B.
Mass spectrometry was performed on an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, San Jose, CA) on line-coupled to the nano-HPLC system. For electrospray ionization, a distal coated SilicaTip (FS-360–50-15-D-20, New Objective, Woburn, MA) and a needle voltage of 1.4 kV were used. The MS method consisted of cycles of one full MS Orbitrap scan (resolution, 60,000; mass range, 300–2000 m/z) and five data-dependent LTQ MS/MS scans (parallel acquisition; 35% collision energy). The dynamic exclusion was set to 30 s. MS/MS data were analyzed with Mascot version 2.1.03 (Matrix Science, Boston, MA) using the following parameters: (i) enzyme, none; (ii) fixed modification, carbamidomethyl (Cys); (iii) variable modification, oxidation (Met); (iv) peptide tolerance, 10 ppm; (v) MS/MS tolerance, 0.8 Da; (vi) peptide charge, 1+, 2+, and 3+; and (vii) instrument, ESI-TRAP. The International Protein Index (IPI) human database (Release 3.70) supplemented with the sequence of the recombinant pro-NGF protein (sequence as provided by the manufacturer without N-terminal signal peptide) was used.
Statistical Analyses
Data analysis and statistics were performed using PRISM 4.0 (GraphPad Software, Inc., San Diego, CA). Statistical analysis was performed using the repeated measures analysis of variance test. Differences between the groups were evaluated with the appropriate post-test (Dunnett). Data shown represent the means ± S.E.
RESULTS
Tryptase, but Not SLIGKV, Increased Amounts of NGF in Cell Culture Supernatant
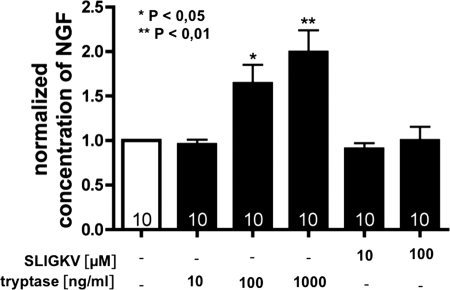
To investigate whether human recombinant β-tryptase can alter the levels of NGF produced by HTPCs and HTPC-Fs within 24 h, NGF levels in cell culture supernatants were determined by a commercial ELISA (n = 10 patients; Fig. 2). Two different concentrations of tryptase (100 and 1000 ng/ml) were effective and in a concentration-dependent manner significantly elevated the levels of NGF. The maximum increase was 1.9-fold over basal secretion, and up to 218 pg of NGF/mg of cellular protein were detected in the supernatants.
FIGURE 2.
NGF levels are increased by tryptase but not by SLIGKV. Using ELISA techniques, we detected that tryptase (100 and 1000 ng/ml) but not the agonist peptide SLIGKV (10 and 100 μm) statistically significantly augmented NGF levels after 24 h. Results are means (bars) plus S.E. are from n = 10 individual HTPC/-F cultures and were normalized to untreated controls. *, p < 0.05; **, p < 0.01.
When the PAR-2 agonist peptide SLIGKV (10 and 100 μm) was tested, levels of NGF in the cell culture media remained unchanged, although 10 μm SLIGKV, like 100 and 1000 ng/ml tryptase, robustly activated PAR-2 as judged from elevated intracellular Ca2+ (not shown). The response of HTPCs and HTPC-Fs to tryptase and SLIGKV did not differ, and hence the data shown do not distinguish between the groups.
Tryptase and SLIGKV Did Not Influence Cell Viability or NGF mRNA Levels
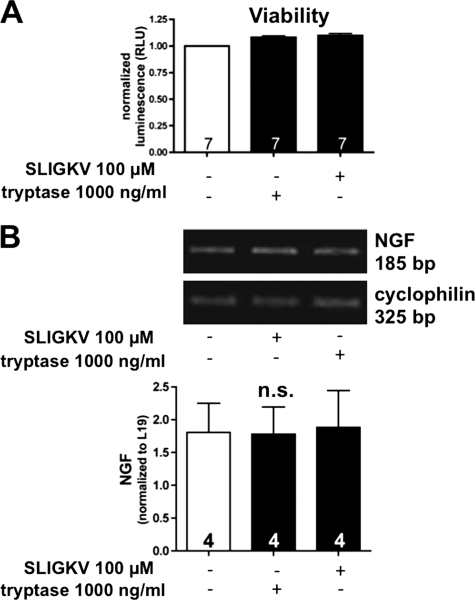
Neither tryptase nor SLIGKV affected cellular ATP levels, a measure of cell viability (n = 7 patients; Fig. 3A). Hence, possible negative effects of SLIGKV that may manifest over a 24-h period can be ruled out.
FIGURE 3.
Tryptase and SLIGKV do not influence cell viability or NGF mRNA levels. A, both tryptase and SLIGKV did not alter cellular ATP levels (n = 7; means ± S.E.; RLU, relative luminescence units). B, semiquantitative RT-PCR (n = 5; top) and quantitative real time PCR (n = 4; bottom) showed that levels of NGF mRNA were not significantly affected by treatment with tryptase or the agonist peptide SLIGKV after 24 h. Bars are means + S.E.; n.s., not significant).
Semiquantitative RT-PCR (n = 5 patients) and qRT-PCR (n = 4 patients) showed that the levels of NGF mRNA were not affected after 24 h of treatment with tryptase (100 ng/ml) or the agonist peptide SLIGKV (100 μm; Fig. 3B). Therefore, NGF does not alter mRNA levels of NGF.
NGF and Its Precursor Pro-NGF in Peritubular Cell Cultures and Supernatant
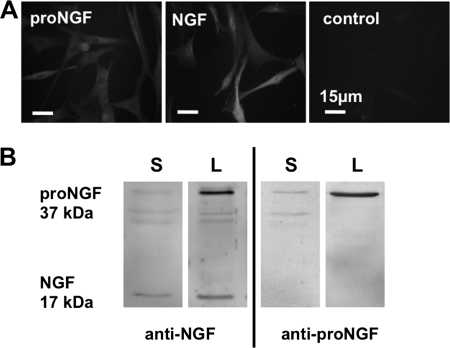
The existence of the proteins NGF and pro-NGF in the cultured cells was confirmed by immunocytochemistry (n = 2; Fig. 4A). The 17-kDa protein NGF and a 37-kDa precursor pro-NGF were detected in lysates of HTPCs and HTPC-Fs (n = 12) and in the conditioned culture medium (n = 6) by Western blotting using two antisera, one recognizing both the pro region of NGF and NGF and another recognizing only mature NGF (Fig. 4B).
FIGURE 4.
NGF and its precursor pro-NGF in human peritubular cells and culture supernatant. A, immunocytochemistry of NGF and pro-NGF in cultured cells (n = 2). B, NGF (17 kDa) and pro-NGF (37 kDa) were detected in lysates of HTPC/-Fs (L; n = 12) and in the conditioned culture medium (S; n = 6) by Western blotting. Note that the NGF antibody recognizes both pro-NGF and NGF, whereas the pro-NGF antibody does not recognize NGF.
Pro-NGF Is a Substrate for Tryptase
Recombinant human β-tryptase (1000 ng/ml tryptase) when added to the cell-free supernatants of HTPCs for 24 h reduced the intensities of immunoreactive pro-NGF bands detected in Western blots by 13–52% when compared with untreated controls (n = 3 experiments; Fig. 5A).
FIGURE 5.
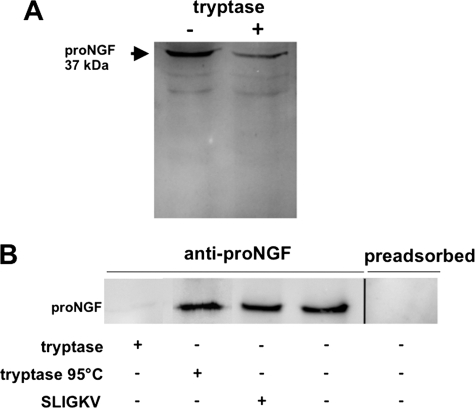
Tryptase can cleave pro-NGF. A, a reduction in the levels of pro-NGF became obvious by Western blot detection of pro-NGF (n = 3) after treatment of the supernatant of HTPCs with or without 1000 ng/ml tryptase for 24 h. B, Western blot studies indicate a decrease of synthetic pro-NGF after treatment with tryptase (1000 ng/ml; 24 h; n = 8) but no decrease by SLIGKV (100 μm; n = 8) or heat-inactivated tryptase (1000 ng/ml; n = 7). Specificity of the pro-NGF antibody was shown by preadsorption (n = 2).
To further investigate the possibility that tryptase can cleave pro-NGF, synthetic pro-NGF instead of conditioned culture medium was used next. Results of Western blots showed that recombinant human β-tryptase (1000 ng/ml; n = 5; Fig. 5B) added for 24 h abolished immunoreactive pro-NGF, a result in line with enzymatic cleavage of pro-NGF by active tryptase. When tryptase was heat-inactivated (1000 ng/ml; n = 4) and when SLIGKV (100 μm; n = 5) was used instead, pro-NGF remained readily detectable. The specificity of the pro-NGF antiserum was verified by preadsorption with the peptide used for immunization (Fig. 5B).
Cleavage Sites of Pro-NGF
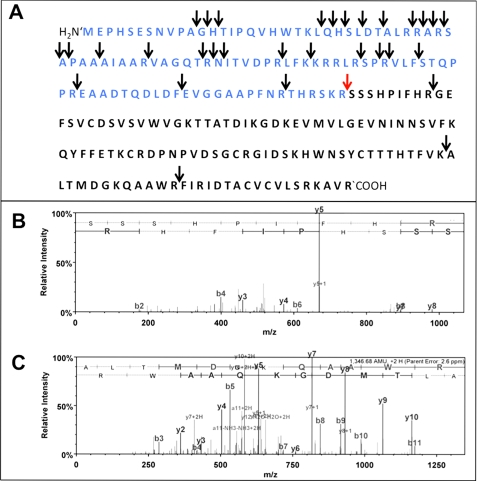
Results from LC-MS/MS demonstrate that under the in vitro conditions used multiple sites within the synthetic pro-NGF and NGF molecules are accessible to cleavage by recombinant tryptase (see Table 1). Fig. 6 shows the cleavage sites of pro-NGF obtained by treatment with recombinant human tryptase (arrows). The pro-NGF sequence contains multiple tryptase cleavage sites, including the site at which furin, proconvertases, and MMP7 are reported to cleave mature NGF (red arrow). This is evident from the detection of peptide SSSHPIFHR (see Table 1 and Fig. 6B). In addition, three cleavage sites within the mature NGF sequence were identified (see Fig. 6).
TABLE 1.
MS/MS analysis of recombinant pro-NGF treated with tryptase
Shown is a list of peptides identified with precursor mass accuracies <5 ppm and Mascot ion scores >25. Arrows indicate tryptase cleavage sites. Start and stop positions refer to the sequence of the recombinant pro-NGF. M(ox), oxidized methionine.
| Ion score | Delta mass | Start | Stop | |
|---|---|---|---|---|
| ppm | ||||
| -↓MEPHSESNVPA↓G | 30 | 1.73 | 1 | 11 |
| -↓MEPHSESNVPAG↓H | 32 | 0.46 | 1 | 12 |
| -↓MEPHSESNVPAGH↓T | 61 | 1.21 | 1 | 13 |
| -↓M(ox)EPHSESNVPAGH↓T | 65 | 1.25 | 1 | 13 |
| H↓TIPQVHWTK↓L | 27 | −0.73 | 14 | 22 |
| K↓LQHSLDT↓A | 50 | 1.59 | 23 | 29 |
| K↓LQHSLDTALR↓R | 60 | −0.01 | 23 | 32 |
| K↓LQHSLDTALRR↓A | 28 | 0.46 | 23 | 33 |
| Q↓HSLDTALR↓R | 36 | −0.35 | 25 | 32 |
| H↓SLDTALR↓R | 49 | 0.74 | 26 | 32 |
| L↓DTALR↓R | 34 | 0.26 | 28 | 32 |
| R↓ARSAPAAAIAAR↓V | 48 | 0.76 | 34 | 45 |
| A↓RSAPAAAIAAR↓V | 57 | 0.91 | 35 | 45 |
| R↓SAPAAAIAAR↓V | 85 | 0.07 | 36 | 45 |
| S↓APAAAIAAR↓V | 48 | −1.16 | 37 | 45 |
| A↓PAAAIAAR↓V | 61 | 1.57 | 38 | 45 |
| A↓AIAAR↓V | 28 | 1.29 | 41 | 45 |
| T↓RNITVDPR↓L | 31 | 1.13 | 51 | 58 |
| R↓NITVDPR↓L | 40 | 0.83 | 52 | 58 |
| R↓NITVDPRLFK↓K | 41 | 0.97 | 52 | 61 |
| N↓ITVDPR↓L | 29 | 0.82 | 53 | 58 |
| R↓SPRVLF↓S | 27 | 1.21 | 67 | 72 |
| P↓RVLFSTQPPR↓E | 35 | 2.22 | 69 | 78 |
| R↓VLFSTQPPR↓E | 72 | −1.17 | 70 | 78 |
| F↓EVGGAAPFNR↓T | 37 | 0.26 | 89 | 98 |
| R↓SSSHPIFHR↓G | 28 | 0.45 | 105 | 113 |
| K↓ALTMDGKQAAWR↓F | 50 | 2.66 | 192 | 203 |
FIGURE 6.
Tryptase cleaves pro-NGF in vitro. A, cleavage sites of tryptase in the pro-NGF sequence (blue) and mature NGF sequence (black) as found by LC-MS/MS analyses in vitro using synthetic human pro-NGF and recombinant human tryptase. Arrows indicate cleavage sites. The cleavage site that is shared by tryptase, furin, and proconvertases is indicated by a red arrow. Note the multiple cleavage sites of the enzyme tryptase within the pro-NGF molecule and the three sites within the mature NGF region. B, MS/MS spectrum of peptide SSSHPIFHR. The N terminus of the peptide represents the N terminus of functional NGF. Annotation of characteristic y and b fragment ions was performed using Scaffold 2.04 software. C, MS/MS spectrum of peptide ALTMDGKQAAWR. The peptide represents an internal fragment of functional NGF. Annotation of characteristic y and b fragment ions was performed using Scaffold 2.04 software.
DISCUSSION
The results of this study show that human β-tryptase can cleave pro-NGF. Consequently, the levels of pro-NGF are lowered, and mature NGF and likely also smaller NGF fragments may result from tryptase action. As all human mast cells contain tryptase, the results imply that active, tryptase-secreting mast cells in the vicinity of pro-NGF/NGF-secreting cells in any human tissue may be able to alter the ratios of pro-NGF/NGF and thus may fundamentally modify the actions of pro-NGF/NGF.
Tryptase is a large tetrameric 135-kDa serine protease contained in all human mast cells (36). The biological functions of tryptase are not fully known but are in part mediated by a specific receptor, PAR-2, a G-protein-coupled receptor activated by tryptase and certain other serine proteases. PAR-2 is expressed and functional in human testicular peritubular cells (37, 38). The enzymatic activation of this receptor can be mimicked by a small peptide analog (SLIGKV) lacking enzymatic activity. Although the initial signaling (i.e. elevations of intracellular Ca2+) can be induced by the agonist peptide and tryptase in human peritubular cells, only tryptase augmented in a concentration-dependent manner the levels of NGF in the cell culture supernatant of HTPC/-Fs. SLIGKV failed to affect NGF levels beyond basal levels. This is not due to possible toxic effects of this peptide as cell viability was not altered. In contrast, the levels of NGF mRNA were affected by neither tryptase nor the analog. Thus, NGF gene expression is not altered by tryptase at least in human peritubular cells. Therefore, enzymatic activity of tryptase not related to PAR-2 activation was tested next. We found in cell-free assays using antibody detection that tryptase, but not heat-inactivated tryptase or the PAR-2 agonist, were effective in abolishing immunoreactive pro-NGF, implying enzymatic cleavage of pro-NGF.
The enzymatic activities of tryptase have been shown not only to catalyze the activation of its receptor (PAR-2) but also to alter the microenvironment around a mast cell by enzymatic cleavage of a number of extracellular proteins (see Introduction), resulting in activation or inactivation of these molecules. The cleavage of proteins occurs in general after arginine or lysine. However, reports from the literature indicate that the preferential cleavage sites of tryptase are characterized by preceding amino acids containing proline at position P4 and by a preference for a positive charged amino acid (e.g. arginine or lysine) at P3 but with little specificity at position P2 (56, 57). The optimal sequence for β-tryptase cleavage is therefore described as P4 Pro, P3 Arg/Lys, P2 X, and P1 Lys/Arg in many of the macromolecular substrates known at least in vitro (56).
If applied to the human pro-NGF sequence, several cleavage sites are possible. One is identical to the natural cleavage site of furin (7, 58). Under in vitro conditions using commercially available synthetic pro-NGF and recombinant tryptase, we found unambiguous experimental evidence for cleavage. Interestingly, cleavage of the synthetic pro-NGF molecule occurred at multiple locations. Tryptase cleavage could be demonstrated at the site of action of furin, proconvertases, and MMP7, which create mature NGF. Finally, cleavage at three sites within the sequence of mature NGF was detected. Taken together, these results show that tryptase degrades pro-NGF and NGF.
Tryptase is a large molecule contained in all human mast cells. It is thought that the tetrameric complex once released from mast cells may be enzymatically active only in the close vicinity of these cells. Mast cells are found in close proximity to NGF-producing cells, e.g. in the testicular peritubular wall but also in other tissues of the human body, including the reproductive tract (59). Changes in the numbers of mast cells associated with human disease have been reported, e.g. in male infertility (33), endometriosis, and polycystic ovarian syndrome (60, 61). Therefore, it must be assumed that the levels and ratios of pro-NGF and NGF are strongly affected by active tryptase-secreting mast cells. Given the preferential affinities of pro-NGF/NGF to receptors linked to cell death or survival, mast cells appear to play an unexpected role in the fate of target cells in health and disease.
Acknowledgments
We thank Daniel Einwang for excellent technical assistance, Marion Adam for help with cell culture and qRT-PCR experiments, and Prof. Dr. F. M. Köhn and Prof. Dr. J. U. Schwarzer for providing human testicular biopsy samples. We gratefully acknowledge the collaboration and help with the qRT-PCR by Prof. Dr. M. Poutanen and Dr. L. Strauss (Turku, Finland).
This work was supported by Deutsche Forschungsgemeinschaft (DFG) Grant MA 1080/16-3 and in part by DFG Grants MA 1080/17-3 and MA 1080/20-1.
- HTPC
- human testicular peritubular cell
- PAR-2
- protease-activated receptor-2
- qRT-PCR
- real time quantitative PCR.
REFERENCES
- 1. Matsuoka I., Meyer M., Thoenen H. (1991) J. Neurosci. 11, 3165–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lipnik-Stangelj M. (2006) Biochem. Pharmacol. 72, 1375–1381 [DOI] [PubMed] [Google Scholar]
- 3. Caroleo M. C., Costa N., Bracci-Laudiero L., Aloe L. (2001) J. Neuroimmunol. 113, 193–201 [DOI] [PubMed] [Google Scholar]
- 4. Furukawa S., Furukawa Y., Satoyoshi E., Hayashi K. (1987) Biochem. Biophys. Res. Commun. 147, 1048–1054 [DOI] [PubMed] [Google Scholar]
- 5. Suter U., Heymach J. V., Jr., Shooter E. M. (1991) EMBO J. 10, 2395–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rattenholl A., Lilie H., Grossmann A., Stern A., Schwarz E., Rudolph R. (2001) Eur. J. Biochem. 268, 3296–3303 [DOI] [PubMed] [Google Scholar]
- 7. Seidah N. G., Benjannet S., Pareek S., Savaria D., Hamelin J., Goulet B., Laliberte J., Lazure C., Chrétien M., Murphy R. A. (1996) Biochem. J. 314, 951–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee R., Kermani P., Teng K. K., Hempstead B. L. (2001) Science 294, 1945–1948 [DOI] [PubMed] [Google Scholar]
- 9. Bruno M. A., Cuello A. C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6735–6740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yoon S. O., Casaccia-Bonnefil P., Carter B., Chao M. V. (1998) J. Neurosci. 18, 3273–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Casaccia-Bonnefil P., Carter B. D., Dobrowsky R. T., Chao M. V. (1996) Nature 383, 716–719 [DOI] [PubMed] [Google Scholar]
- 12. Arévalo J. C., Wu S. H. (2006) Cell. Mol. Life Sci. 63, 1523–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nykjaer A., Lee R., Teng K. K., Jansen P., Madsen P., Nielsen M. S., Jacobsen C., Kliemannel M., Schwarz E., Willnow T. E., Hempstead B. L., Petersen C. M. (2004) Nature 427, 843–848 [DOI] [PubMed] [Google Scholar]
- 14. Paiardini A., Caputo V. (2008) Neuropeptides 42, 205–214 [DOI] [PubMed] [Google Scholar]
- 15. Clewes O., Fahey M. S., Tyler S. J., Watson J. J., Seok H., Catania C., Cho K., Dawbarn D., Allen S. J. (2008) J. Neurochem. 107, 1124–1135 [DOI] [PubMed] [Google Scholar]
- 16. Fahnestock M., Yu G., Coughlin M. D. (2004) Prog. Brain Res. 146, 101–110 [DOI] [PubMed] [Google Scholar]
- 17. Reinshagen M., Geerling I., Eysselein V. E., Adler G., Huff K. R., Moore G. P., Lakshmanan J. (2000) J. Neurochem. 74, 2127–2133 [DOI] [PubMed] [Google Scholar]
- 18. Dissen G. A., Hirshfield A. N., Malamed S., Ojeda S. R. (1995) Endocrinology 136, 4681–4692 [DOI] [PubMed] [Google Scholar]
- 19. Mayerhofer A., Dissen G. A., Parrott J. A., Hill D. F., Mayerhofer D., Garfield R. E., Costa M. E., Skinner M. K., Ojeda S. R. (1996) Endocrinology 137, 5662–5670 [DOI] [PubMed] [Google Scholar]
- 20. Julio-Pieper M., Lozada P., Tapia V., Vega M., Miranda C., Vantman D., Ojeda S. R., Romero C. (2009) J. Clin. Endocrinol. Metab. 94, 3065–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Labouyrie E., Dubus P., Groppi A., Mahon F. X., Ferrer J., Parrens M., Reiffers J., de Mascarel A., Merlio J. P. (1999) Am. J. Pathol. 154, 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yardley G., Relf B., Lakshmanan J., Reinshagen M., Moore G. P. (2000) Exp. Dermatol. 9, 283–289 [DOI] [PubMed] [Google Scholar]
- 23. Kendall T. J., Hennedige S., Aucott R. L., Hartland S. N., Vernon M. A., Benyon R. C., Iredale J. P. (2009) Hepatology 49, 901–910 [DOI] [PubMed] [Google Scholar]
- 24. Girard B. M., Malley S. E., Vizzard M. A. (2011) Am. J. Physiol. Renal Physiol. 300, F345–F355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chakrabarti S., Sima A. A., Lee J., Brachet P., Dicou E. (1990) Brain Res. 523, 11–15 [DOI] [PubMed] [Google Scholar]
- 26. Delsite R., Djakiew D. (1999) Prostate 41, 39–48 [DOI] [PubMed] [Google Scholar]
- 27. Hayashi K., Storesund T., Schreurs O., Khuu C., Husvik C., Karatsaidis A., Helgeland K., Martin-Zanca D., Schenck K. (2007) Eur. J. Oral Sci. 115, 344–354 [DOI] [PubMed] [Google Scholar]
- 28. Chen Y., Dicou E., Djakiew D. (1997) Mol. Cell. Endocrinol. 127, 129–136 [DOI] [PubMed] [Google Scholar]
- 29. Aloe L. (2001) Arch. Physiol. Biochem. 109, 354–356 [DOI] [PubMed] [Google Scholar]
- 30. Welker P., Grabbe J., Gibbs B., Zuberbier T., Henz B. M. (2000) Immunology 99, 418–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schell C., Albrecht M., Mayer C., Schwarzer J. U., Frungieri M. B., Mayerhofer A. (2008) Endocrinology 149, 1678–1686 [DOI] [PubMed] [Google Scholar]
- 32. Spinnler K., Köhn F. M., Schwarzer U., Mayerhofer A. (2010) Hum. Reprod. 25, 2181–2187 [DOI] [PubMed] [Google Scholar]
- 33. Meineke V., Frungieri M. B., Jessberger B., Vogt H., Mayerhofer A. (2000) Fertil. Steril. 74, 239–244 [DOI] [PubMed] [Google Scholar]
- 34. Frungieri M. B., Weidinger S., Meineke V., Köhn F. M., Mayerhofer A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15072–15077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Frungieri M. B., Calandra R. S., Lustig L., Meineke V., Köhn F. M., Vogt H. J., Mayerhofer A. (2002) Fertil. Steril. 78, 298–306 [DOI] [PubMed] [Google Scholar]
- 36. Schwartz L. B., Irani A. M., Roller K., Castells M. C., Schechter N. M. (1987) J. Immunol. 138, 2611–2615 [PubMed] [Google Scholar]
- 37. Adam M., Schwarzer J. U., Köhn F. M., Strauss L., Poutanen M., Mayerhofer A. (2011) Hum. Reprod., in press [DOI] [PubMed] [Google Scholar]
- 38. Albrecht M., Rämsch R., Köhn F. M., Schwarzer J. U., Mayerhofer A. (2006) J. Clin. Endocrinol. Metab. 91, 1956–1960 [DOI] [PubMed] [Google Scholar]
- 39. Molino M., Barnathan E. S., Numerof R., Clark J., Dreyer M., Cumashi A., Hoxie J. A., Schechter N., Woolkalis M., Brass L. F. (1997) J. Biol. Chem. 272, 4043–4049 [DOI] [PubMed] [Google Scholar]
- 40. Schwartz L. B., Bradford T. R., Littman B. H., Wintroub B. U. (1985) J. Immunol. 135, 2762–2767 [PubMed] [Google Scholar]
- 41. Gruber B. L., Marchese M. J., Suzuki K., Schwartz L. B., Okada Y., Nagase H., Ramamurthy N. S. (1989) J. Clin. Investig. 84, 1657–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hallgren J., Spillmann D., Pejler G. (2001) J. Biol. Chem. 276, 42774–42781 [DOI] [PubMed] [Google Scholar]
- 43. Lohi J., Harvima I., Keski-Oja J. (1992) J. Cell. Biochem. 50, 337–349 [DOI] [PubMed] [Google Scholar]
- 44. Tam E. K., Caughey G. H. (1990) Am. J. Respir. Cell Mol. Biol. 3, 27–32 [DOI] [PubMed] [Google Scholar]
- 45. Caughey G. H., Leidig F., Viro N. F., Nadel J. A. (1988) J. Pharmacol. Exp. Ther. 244, 133–137 [PubMed] [Google Scholar]
- 46. Kaminska R., Helisalmi P., Harvima R. J., Naukkarinen A., Horsmanheimo M., Harvima I. T. (1999) J. Invest. Dermatol. 113, 567–573 [DOI] [PubMed] [Google Scholar]
- 47. Fajardo I., Pejler G. (2003) Biochem. J. 369, 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Imamura T., Dubin A., Moore W., Tanaka R., Travis J. (1996) Lab. Invest. 74, 861–870 [PubMed] [Google Scholar]
- 49. Rauter I., Krauth M. T., Westritschnig K., Horak F., Flicker S., Gieras A., Repa A., Balic N., Spitzauer S., Huss-Marp J., Brockow K., Darsow U., Behrendt H., Ring J., Kricek F., Valent P., Valenta R. (2008) J. Allergy Clin. Immunol. 121, 197–202 [DOI] [PubMed] [Google Scholar]
- 50. Mayerhofer A., Frungieri M. B., Fritz S., Bulling A., Jessberger B., Vogt H. J. (1999) J. Androl. 20, 341–347 [PubMed] [Google Scholar]
- 51. Schell C., Albrecht M., Spillner S., Mayer C., Kunz L., Köhn F. M., Schwarzer U., Mayerhofer A. (2010) Endocrinology 151, 1257–1268 [DOI] [PubMed] [Google Scholar]
- 52. Li X., Strauss L., Kaatrasalo A., Mayerhofer A., Huhtaniemi I., Santti R., Mäkelä S., Poutanen M. (2006) Endocrinology 147, 1271–1277 [DOI] [PubMed] [Google Scholar]
- 53. Strauss L., Kallio J., Desai N., Pakarinen P., Miettinen T., Gylling H., Albrecht M., Mäkelä S., Mayerhofer A., Poutanen M. (2009) Endocrinology 150, 2865–2872 [DOI] [PubMed] [Google Scholar]
- 54. Rey-Ares V., Lazarov N., Berg D., Berg U., Kunz L., Mayerhofer A. (2007) Reprod. Biol. Endocrinol. 5, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Saller S., Kunz L., Dissen G. A., Stouffer R., Ojeda S. R., Berg D., Berg U., Mayerhofer A. (2010) Hum. Reprod. 25, 969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Harris J. L., Niles A., Burdick K., Maffitt M., Backes B. J., Ellman J. A., Kuntz I., Haak-Frendscho M., Craik C. S. (2001) J. Biol. Chem. 276, 34941–34947 [DOI] [PubMed] [Google Scholar]
- 57. Hallgren J., Pejler G. (2006) FEBS J. 273, 1871–1895 [DOI] [PubMed] [Google Scholar]
- 58. Pagadala P. C., Dvorak L. A., Neet K. E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17939–17943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bose C. K. (2009) J. Stem Cells 4, 217–227 [PubMed] [Google Scholar]
- 60. Anaf V., Chapron C., El Nakadi I., De Moor V., Simonart T., Noël J. C. (2006) Fertil. Steril. 86, 1336–1343 [DOI] [PubMed] [Google Scholar]
- 61. Heider U., Pedal I., Spanel-Borowski K. (2001) Fertil. Steril. 75, 1141–1147 [DOI] [PubMed] [Google Scholar]