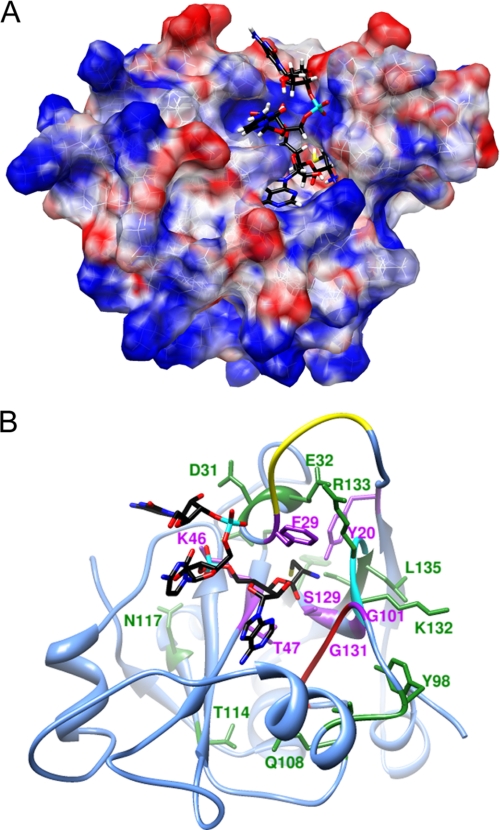
FIGURE 2.
Computational model of H. influenzae YbaK·CCA-Cys complex. A. The electrostatic potential surface of YbaK was generated by APBS software (65). The cysteine moiety of the substrate (yellow) is located in a deep cleft in the active site. B, ribbon representation of YbaK structure with CCA-Cys substrate shown primarily in black. The NNQHF loop, GXXXP loop, and KRG loop are highlighted in yellow, red, and cyan, respectively. Residues changed in this study are highlighted in purple (deacylation rate decreased >5-fold upon mutagenesis) or green (deacylation rate affected <3-fold upon mutagenesis).