Abstract
The RecA-dependent DNA damage response pathway (SOS response) appears to be the major DNA repair mechanism in most bacteria, but it has been suggested that a RecA-independent mechanism is responsible for controlling expression of most damage-inducible DNA repair genes in Mycobacterium tuberculosis. The specific reparative responses and molecular mediators involved in the DNA repair mechanism remain largely unclear in this pathogen and its related species. In this study, a mycobacterial ClpR-like regulator, corresponding to Rv2745c in M. tuberculosis and to Ms2694 in M. smegmatis mc2155, was found to interact with the promoter regions of multiple damage-inducible DNA repair genes. Specific binding of the ClpR-like factor to the conserved RecA-independent promoter RecA-NDp motif was then confirmed using in vitro electrophoretic mobility shift assays as well as in vivo chromatin immunoprecipitation experiments. The ClpR knock-out experiments, in combination with quantitative real time PCR assays, demonstrated that the expression of these RecA-independent genes were significantly down-regulated in the mutant strain of M. smegmatis in response to a DNA-damaging agent compared with the wild type strain. Furthermore, the ClpR-like factor was shown to contribute to mycobacterial genomic stability. These results enhance our understanding of the function of the ClpR regulator and the regulatory mechanism of RecA-independent DNA repair in mycobacteria.
Keywords: Bacterial Transcription, DNA-binding Protein, DNA-Protein Interaction, Gene Regulation, Promoters, Protein DNA-Interaction, Transcription Factors, RecA, Mycobacteria
Introduction
Mycobacterium tuberculosis, the causative agent of tuberculosis, infects nearly one-third of the world population and kills ∼2 million people annually (1). As an intracellular pathogen, M. tuberculosis must confront a variety of DNA-damaging assaults during infection such as antimicrobial reactive oxygen intermediates and reactive nitrogen intermediates generated by the host (2). M. tuberculosis has the ability to respond to DNA damage and overcome gene mutations for survival, but the mechanisms of the DNA damage response are not well understood (3).
Two different mechanisms, RecA/LexA-dependent and RecA-independent, have been described for DNA damage repair in the SOS response. In most bacteria, the SOS response depends on the RecA/LexA system and constitutes transcriptional regulation in response to DNA damage (4). In the classical RecA/LexA-dependent mechanism, RecA coordinates the expression of reparative genes in response to DNA damage, whereas LexA is a repressor protein and binds to the SOS boxes of other promoters to inhibit transcription. The RecA protein can stimulate LexA autocatalytic cleavage and alleviate the repression by LexA to increase the expression of DNA repair genes after DNA damage. Expression of DNA repair genes and recA are suppressed by the LexA repressor under normal growth conditions. Following DNA damage, RecA, in conjunction with LexA, controls the expression of a set of DNA repair and cell division genes to promote bacterial survival (5).
The majority of inducible DNA repair genes in M. tuberculosis show RecA-independent activation (6–8), although the RecA/LexA-dependent mechanism has also been reported in this human pathogen (9). Transcription of the M. tuberculosis recA gene can be activated by two different promoters. Both transcription pathways are induced by DNA damage but only one is regulated by the RecA/LexA mechanism (9). This suggests that an unknown RecA-independent mechanism for DNA damage repair exists in M. tuberculosis. However, little is known about this RecA-independent mechanism for recA expression, previously reported only in Acinetobacter calcoaceticus (10). Therefore, although RecA-dependent regulation appears to be a major mechanism for the DNA damage response in most bacteria, a RecA-independent mechanism may be responsible for regulating expression of damage-inducible DNA repair genes in the human pathogen M. tuberculosis (6–9, 11).
Recently, a highly conserved motif was characterized by bioinformatics within the promoter regions of 47 RecA-independent genes in M. tuberculosis (12). The identified motif had high conservation in two blocks of consensus sequences, tTGTC(G/A)gtg and TAnnnT, which are similar to σ70 recognition elements except that the two motifs in the RecA-independent gene promoters are positioned only eight nucleotides apart (12). This RecA-independent promoter (RecA-NDp) has been associated with DNA repair genes in M. tuberculosis and in many other Actinomycetales (12). However, the transcription factor that recognizes the motif remains to be characterized in M. tuberculosis and related species.
The clp gene regulator ClgR acts as transcriptional activator of the clp protease, and orthologs (termed ClpRs) have been found in all mycobacteria, for example Rv2745c in M. tuberculosis, and in some other actinomycetes (13–15). A transcriptome analysis implicated that ClpR not only controls transcription of clp protease but also controls genes involved in DNA repair in Corynebacterium glutamicum (16). Recent studies have also found that Rv2745c is involved in the regulation of clp genes in M. tuberculosis (17, 18). However, the correlation between Rv2745c and its mycobacteria homologs in the expression of DNA damage-inducible repair genes remains largely unclear.
An important role for transcriptional regulation during the infection of M. tuberculosis is suggested by the presence of genes encoding >140 putative regulatory proteins as well as 13 RNA polymerase σ-factors in the M. tuberculosis genome (19, 20). In this study, the ClpR-like regulator Rv2745c of M. tuberculosis and Ms2694 in M. smegmatis were shown to bind specifically to the conserved RecA-NDp motif in mycobacteria. We show that the regulator could play an essential role in inducing expression of these DNA repair genes in response to DNA damage. These findings are the first detailed description of the RecA/LexA-independent DNA repair mechanism in mycobacteria.
EXPERIMENTAL PROCEDURES
Strains, Enzymes, Primers and Plasmids
Escherichia coli DH5α strains (Stratagene) were used for cloning, whereas E. coli BL21 (DE3) cells (Novagen) were used as the host strain to express recombinant proteins. The pBT and pTRG vectors and E. coli XR host strain were purchased from Stratagene. The pGEX-4T-1 vector was purchased from GE Healthcare. Restriction enzymes, T4 ligase, Pyrobest DNA polymerase, modification enzymes, and all antibiotics were obtained from TaKaRa Bio, Inc.. All reagents for the one-hybrid assay were purchased from Stratagene. All PCR primers were synthesized by Invitrogen (supplemental Table S1). Nickel-nitrilotriacetic acid-agarose was obtained from Qiagen.
Bacterial One-hybrid Assay
About 150 putative transcription regulatory genes were cloned into the pTRG vector. A sub-genomic library containing 150 putative transcription factor genes of M. tuberculosis was produced by mixing these recombinant plasmids as described previously (21). The promoters of the M. tuberculosis and M. smegmatis genes were amplified using their primers (supplemental Table S1) and were then inserted directly into the XcmI site of pBXcmT. A pair of pBX/pTRG plasmids was co-transformed into the reporter strain as a negative control, and its growth was then tested, together with the self-activation control on the selective medium containing 3-amino-1,2,4-triazole, Kanr, Strr, and Chlr. Positive growth co-transformants were selected on a screening medium plate containing 20 mm 3-amino-1,2,4-triazole, 16 μg/ml streptomycin, 15 μg/ml tetracycline, 34 μg/ml chloramphenicol, and 50 μg/ml kanamycin. The plates were incubated at 30 °C for 3–4 days. CK+ refers to co-transformant containing pRomoter-Rv2031p and pTRG-Rv3133c as a positive control (21). CK− refers to co-transformant containing pRomoter-Rv2031p and pTRG-Rv3133c-deltaC as a negative control (21).
Protein Expression and Purification
Rv2745c, Ms2694, and their mutant genes were amplified from genomic DNA using their primers (supplemental Table S1) and were cloned into the pGEX-4T-1 vector to produce recombinant plasmids (supplemental Table S2). E. coli BL21 (DE3) cells transformed with the recombinant plasmids were grown at 37 °C in 1 liter of LB medium containing 100 μg/ml ampicillin up to an optical density of 0.9 at 600 nm. Protein expression was induced at 16 °C for 20 h by the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside. Harvested cells were resuspended and sonicated. The cell lysate was centrifuged at 10,000 × g for 30 min, and the clear supernatant was loaded on to a GST affinity purification column. The column-bound protein was washed with PBS buffer. The protein was then eluted using PBS buffer containing 10 mm reduced glutathione (GSH). The elution was then dialyzed overnight at 4 °C and stored at −80 °C until needed. Purified proteins were resolved by 12% SDS-PAGE. Protein concentration was detected by the Bradford method.
EMSA
DNA probes for gel shift assays were obtained by PCR amplification from M. tuberculosis and M. smegmatis genomic DNA. Amplification products were purified with BioFlux PCR DNA Purification kit (BioFlux). Purified DNA fragments were labeled with [γ-32P]ATP using T4 polynucleotide kinase (Takara) following the manufacturer's instructions. 32P-labeled DNA fragments were incubated at room temperature for 30 min with various amounts of proteins in a total volume of 15 μl of an EMSA buffer consisting of 50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 1 mm DTT, and 50 mm NaCl. The protein-DNA mixtures were directly subjected to 5% native PAGE containing 0.5×Tris borate-EDTA (TBE) buffer. Electrophoresis was performed at 150 V at 4 °C in an ice-cold bath until bromphenol blue reached the bottom of the gel. Gels were exposed to a storage phosphor screen overnight at room temperature. The images were acquired by Typhoon Scanner (GE Healthcare).
Construction of Ms2694 Deletion Mutant of M. smegmatis mc2155
Knock-out of the Ms2694 gene from M. smegmatis mc2155 was performed. A suicide plasmid pMind carrying a hygromycin resistance gene was constructed, and a sacB gene was inserted to confer sensitivity to sucrose as a negative selection marker. The recombinant plasmid pMind2694 was electroporated into M. smegmatis mc2155 and selected on LB medium containing 100 μg/ml hygromycin and 2% sucrose. Genomic DNA from allelic-exchange mutants in which the Ms2694 gene had been deleted was identified by restriction digestion and confirmed by PCR analysis using primers on each side of the Ms2694 and the hygromycin gene.
Preparation of Gene Probes and Southern Blot Assays
A 317-bp probe corresponding to the sequence of the Ms2694 upstream genomic fragment of M. smegmatis was obtained by PCR using the primer pair 5′-AGGTGCTCGACGCTGTCGGT-3′ and 5′-GTGGGATGTCTTACCGCAGC-3′). The PCR product was labeled with digoxigenin dUTP (Roche Applied Science) and was used to detect the size change of the MluI-digested genomic fragment of M. smegmatis before and after recombination. Total DNA of M. smegmatis or M. smegmatis ΔMs2694 was digested completely using MluI, and the resulting fragments were fractionated by agarose gel electrophoresis (1%), transferred to a nylon membrane, and hybridized with the 317-bp probe. Southern blotting and DNA hybridization were performed according to the manufacturer's instructions (Roche Applied Science).
Scanning Electron Microscopy Observation
M. smegmatis cells prepared for SEM observation were grown and harvested by centrifugation. The bacterial pellets were resuspended and incubated at 4 °C for 24 h in 2.5% glutaraldehyde solution. The cells were washed and then dehydrated by sequential 15-min treatments in 30, 50, 75, 85, 95, and 100% ethanol. The incubation in 100% ethanol was repeated to ensure complete dehydration. Samples were dried up to the critical point, sputter-coated with gold, and observed using a scanning electron microscope (S570; Hitachi, Tokyo, Japan). The p values of the cell length data were calculated by unpaired two-tailed Student's t test using GraphPad Prism (version 5).
Quantitative Real-time PCR Assay
Isolation of mRNA and cDNA preparation of wild type and ΔMs2694 M. smegmatis strains were performed as described previously (22). For real-time PCR analysis, gene-specific primers (supplemental Table S1) were used, and first-strand cDNAs were synthesized using a ReverTra Ace first-strand cDNA synthesis kit (TOYOBO) and SuperScript II reverse transcriptase (Invitrogen), according to the manufacturer's instructions. Each PCR reaction (20 μl) contained 10 μl of 2× SYBR Green Master Mix Reagent (Applied Biosystems), 1.0 μl of cDNA samples, and 200 nm primers. The reactions were performed in a Bio-Rad IQ5 real-time PCR machine. The thermocycling conditions were 95 °C for 5 min and 40 cycles at 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. Amplification specificity was assessed using melting curve analysis. Relative expression levels were normalized using sigA as an invariant transcript (internal control), and data were analyzed using the ΔΔCT method as described previously (23). The average relative expression levels and S.D. were determined from three independent experiments. For a positive control, the total genomic DNA of each strain was used as a template to amplify a product. The cDNA of a recombinant strain containing empty pMind was used as a template for a negative control.
ChIP Assay
The in vivo interactions between Ms2694 and the promoters of M. smegmatis DNA repair genes were investigated by ChIP assays. Exponentially growing M. smegmatis cells were fixed with 1% formaldehyde at room temperature for 10 min, and the reaction was stopped by adding glycine (final concentration 0.125 m). Cross-linked cells were harvested and resuspended in 1 ml of TBSTT (TBS + 0.2% Triton X-100 + 0.05% Tween-20). The sample was sonicated on ice, and the average DNA fragment size was determined to be ∼0.5 kb. A 100-μl sample of the extract was saved as the input fraction. The remaining 900 μl was then incubated with 10 μl of antibodies against Ms2694 under rotation for 3 h at 4 °C. The complexes were immunoprecipitated with 20 μl 50% protein A-agarose for 1 h under rotation at 4 °C. The immunocomplex was recovered by centrifugation and resuspended in 100 μl TE (20 mm Tris-HCl, pH 7.8, 10 mm EDTA, 0.5% SDS). Cross-linking was reversed for 6 h at 65 °C. The DNA samples of the input and ChIP were purified, resuspended in 50 μl TE and analyzed by PCR. Each experiment was performed in duplicate and repeated twice. The amplification protocol included one denaturation step of 5 min at 95 °C and then 32 cycles of 1 min at 95 °C, 1 min at 60 °C, and 1 min at 72 °C.
Estimation of Mutation Frequencies and Rates
The mutation frequency of the streptomycin-resistant gene in the wild type strain and ΔMs2694 mutant strain were analyzed according to a previously published procedure (24). Briefly, single colonies from M. smegmatis strains were used to generate the inocula, which were grown in Middlebrook 7H9 medium (Difco) supplemented with 10% (v/v) albumin-dextrose-catalase (Merck), 0.2% glycerol, 0.1% Tween 80 to an optical density at 600 nm of 1.2∼1.5. Cell suspensions of M. smegmatis were harvested and passed through a sterile filter (5 μm) before dilution to ensure a single cell suspension. Then, 1 ml of the cultures were plated onto 7H10 solid medium containing streptomycin (50 μg/ml) in triplicate to monitor spontaneous mutation frequencies. The remaining cultures were diluted to 10−6, and 200 μl of the dilution were plated on 7H10 antibiotic-free solid medium in triplicate for cfu determination. Mutation frequencies were calculated as reported previously (24).
Rates of spontaneous mutation of M. smegmatis strains to streptomycin resistance were determined by Luria-Delbrück fluctuation analysis (25) using the method described by Machowski et al. (26) with a little modification. Single colonies from M. smegmatis wild type strain, Ms2694 deletion strain, and the Ms2694 deletion strain complemented with a Ms2694 expression plasmid pMindD2694 were used to generate the inocula, which were grown in Middlebrook 7H9 medium (Difco) supplemented with 10% (v/v) albumin-dextrose-catalase (Merck), 0.2% glycerol, 0.1% Tween 80 to an optical density at 600 nm of 1.2∼1.5. Additional kanamycin (50 μg/ml) was added in the medium for the stabilization of pMindD2694 in the complemented mutant strain. The inocula were diluted to a cell density of ∼103 cfu/ml and 0.5-ml aliquots were grown at 37 °C in 5-ml culture tubes for 3 days, with shaking at 180 rpm. To ensure that no pre-existing Strr mutants were present after expansion from the colonies and dilution to seeding density, inocula were plated directly onto medium containing streptomycin (12.5 μg/ml). The final number of cells in the culture (Nt) and the observed number of mutant in the same culture (r) were determined by plating without and with streptomycin, respectively. For determination of r values, the entire contents of a culture tube were plated on streptomycin supplemented medium following the removal of a 100-μl aliquot for cfu determination. For determination of Nt values, the 100-μl cultures were diluted to 10−7 and plated on the 7H10 solid medium without streptomycin. cfu were scored after 3 days of growth. Mutation rates were calculated by the method of the median (27) using the formula: mutation rate = m/Nt, where m was calculated from r via the Lea-Coulson equation: r/m − ln(m) − 1.24 = 0 using the web tool FALCOR (Fluctuation Analysis Calculator) (28). p values of the results were calculated by unpaired two-tailed Student's t test using GraphPad Prism (version 5).
RESULTS
ClpR-like Regulator of M. tuberculosis, Rv2745c, Binds Specifically to RecA-NDp Motif
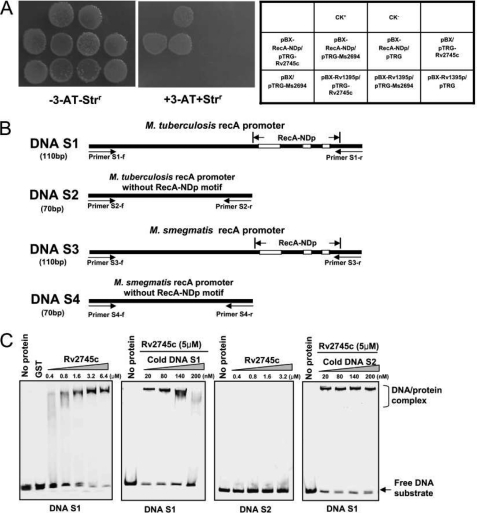
To identify the as yet unknown regulator that binds with the conserved RecA-NDp motif, we first screened the transcriptional regulator library of M. tuberculosis (21), focusing on the regulatory sequences of 20 previously reported genes containing the RecA-NDp motif in their promoter regions (14). Using a one-hybrid technique (21), we isolated a ClpR-like regulator of M. tuberculosis, Rv2745c, which could interact with the regulatory sequences of these genes. As shown in Fig. 1A, the co-transformants containing pTRG-Rv2745c/pBX-RecA-NDp grew well on the screening medium. In contrast, no growth was observed for their self-activated controls or for the co-transformant with pTRG-Rv2745c and an unrelated mycobacterial gene promoter cloned in pBX-Rv1395p that lacked the RecA-NDp motif. In addition, positive controls (CK+) also grew well on the medium, whereas negative controls (CK−) were incapable of growth on the same screening medium (Fig. 1A). These results suggested that the selective system we used worked well and that Rv2745c could interact with the conserved RecA-NDp motif in M. tuberculosis.
FIGURE 1.
Bacterial one-hybrid and EMSA assays for binding of Rv2745c specifically with the RecA-NDp motif. A, bacterial one-hybrid assays. The promoter of the M. tuberculosis recA gene containing the RecA-NDp motif was cloned into pBXcmT (21). Rv2745c and Ms2694 in M. smegmatis mc2155 were cloned into pTRG vectors, respectively. The E. coli XL1-Blue MRF′ Kan strain was used for the routine propagation of all pBXcmT and pTRG recombinant plasmids. A pair of pBXcmT/pTRG plasmids was co-transformed into the reporter strain, and then its growth was tested together with the self-activation controls on a selective medium containing 3-amino-1,2,4-triazole (3-AT), Kanr, Strr, and Chlr as described under “Experimental Procedures.” An outline of the plates is shown in the right panel. Each unit represents the corresponding co-transformant in the plates. The Rv1395 promoter does not contain the RecA-NDp motif and was used as an additional negative control to confirm the specificity of the binding of Rv2745c or Ms2694 to the RecA-NDp motif. B, DNA substrates designed for EMSA assays. Four DNA substrates were amplified from M. tuberculosis and M. smegmatis mc2155 genomic DNA. Both substrate S1 and S3 contain the RecA-NDp. S2 and S4 do not contain the conserved motif. Primers for amplifying the DNA substrates and the location of the RecA-NDp motif are indicated. C, EMSA assays for the binding of Rv2745c with the DNA substrate with or without the RecA-NDp motif. Rv2745c bound to DNA S1, but not DNA S2. Unlabeled cold DNA substrates were used to compete with the [γ-32P]ATP-labeled DNA. GST was used as a negative control. Cold DNA S1, but not S2, could competitively inhibit the binding of Rv2745c to the labeled DNA substrate S1.
An EMSA assay confirmed the specificity of Rv2745c binding to the RecA-NDp motif. We designed two DNA substrates, S1 with the conserved motif and S2 without the conserved motif, and amplified them from M. tuberculosis genomic DNA (Fig. 1B). As shown in Fig. 1C, Rv2745c strongly bound to DNA S1 as evidence by the protein-DNA complex that appeared on the gel as the concentration of Rv2745c increased stepwise from 0.4 to 6.4 μm (Fig. 1C, left panel). In contrast, no complex was observed for DNA S2 as the concentration of Rv2745c increased. In a competition EMSA experiment, the amount of the Rv2745c protein-DNA S1 complex decreased as more unlabeled cold DNA S1 (from 20 nm to 200 nm) was added to the reactions. This indicated that cold DNA S1 could competitively inhibit the binding of Rv2745c to the labeled DNA substrate. In contrast, DNA S2, which did not have the conserved motif, did not compete with Rv2745c binding to the labeled DNA substrate (Fig. 1C, right panel), strongly suggesting that Rv2745c could bind specifically to the RecA-NDp motif.
ClpR-like Regulator of M. smegmatis, Ms2694, Interacts with RecA-NDp Motif Both in Vitro and in Vivo
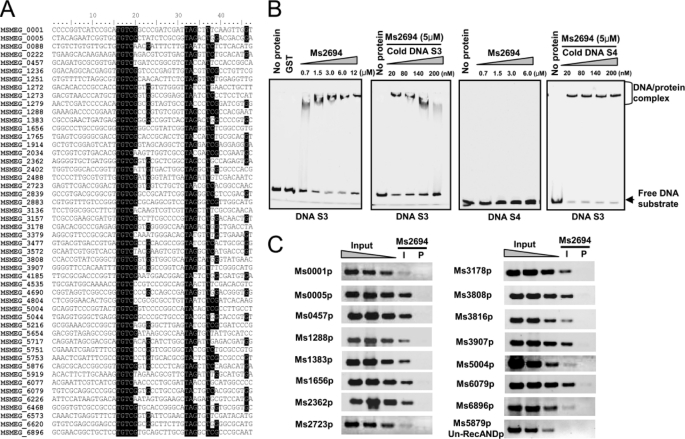
In the bacterial one-hybrid assays (Fig. 1A), the Rv2745c homolog Ms2694 and the ClpR-like regulator of M. smegmatis also interacted with the RecA-NDp motif (Fig. 1A). In a BLAST search, a conserved RecA-NDp motif was also found within the regulatory regions of 49 M. smegmatis genes, of which 15 genes were annotated as being involved in DNA repair (Fig. 2A). To assay the specificity of Ms2694 binding, we designed a pair of DNA substrates, S3 and S4, and amplified them from M. smegmatis genomic DNA. The S3 fragment contained the conserved RecA-NDp motif, whereas S4 did not (Fig. 1B). Similar to Rv2745c, Ms2694 exhibited concentration-dependent DNA binding activity to DNA S3, but not to DNA S4, in EMSA assays (Fig. 2B). In addition, cold DNA S3 could competitively inhibit the binding of Ms2694 with the labeled DNA substrate, whereas cold DNA S4 had no effect on Ms2694 binding (Fig. 2B, right panel).
FIGURE 2.
In vivo and in vitro assays for binding of the M. smegmatis Ms2694 with the RecA-NDp motif. A, BLAST search for the conserved RecA-NDp motif and the DNA repair genes in M. smegmatis. Positions conserved in the sequences of the M. smegmatis genes are shaded. B, EMSA assays for the binding of Ms2694 with the DNA substrates S3 and S4. Ms2694 bound to DNA S3, but not DNA S4. The unlabeled cold DNA substrates were used to compete with the [γ-32P]ATP-labeled DNA. GST was used as a negative control. Cold DNA S3, but not S4, could competitively inhibit the binding of Ms2694 to the labeled DNA substrate S3. Free DNA substrate and DNA-protein complex are indicated. C, ChIP assays for the binding of Ms2694 with the RecA-independent promoter motif. Chromatin immunoprecipitation using preimmune (P) or immune (I) sera raised against Ms2694. Exponentially growing M. smegmatis cells were fixed with 1% formaldehyde. Cross-linked cells were resuspended and sonicated on ice. A 100-μl sample of the extract was saved as the input fraction. The remaining 900 μl was then incubated with 10 μl of antibodies against Ms2694 at 4 °C. The complexes were immunoprecipitated with 20 μl 50% protein A-agarose. The immunocomplex was recovered by centrifugation and resuspended in 100 μl of TE. Cross-linking was reversed for 6 h at 65 °C. The DNA samples of the input and ChIP were purified and resuspended in 50 μl of TE. DNA recovered from immunoprecipitates was amplified with primers specific for DNA repair genes or to an unrelated mycobacterial promoter of Ms5879, which was used as a negative control.
The physiological significance of the in vitro EMSA assays was investigated with ChIP experiments. Fifteen DNA repair genes, including recA (Ms2723), recB (Ms3907), dnaN (Ms0001), ligA (Ms2362), uvrA (Ms3808), uvrB (Ms3816), dnaE (Ms3178), and radA (Ms6079), were selected for in vivo DNA-binding assays. As shown in Fig. 2C, the interaction of Ms2694 with the promoters of these DNA repair genes were validated by the observations that the specific target DNA could be amplified from immunoprecipitates pulled down using the Ms2694 antibody (Fig. 2C). In contrast, the Ms2694 antibody could not immunoprecipitate the DNA from the promoter region of a negative control gene (Ms5879) that did not contain the conserved RecA-NDp motif. These results indicate that Ms2694 can interact specifically with the RecA-NDp motif in vivo in M. smegmatis.
ClpR-like Factor of M. smegmatis Regulates Expression of RecA-independent Genes in Response to DNA Damage
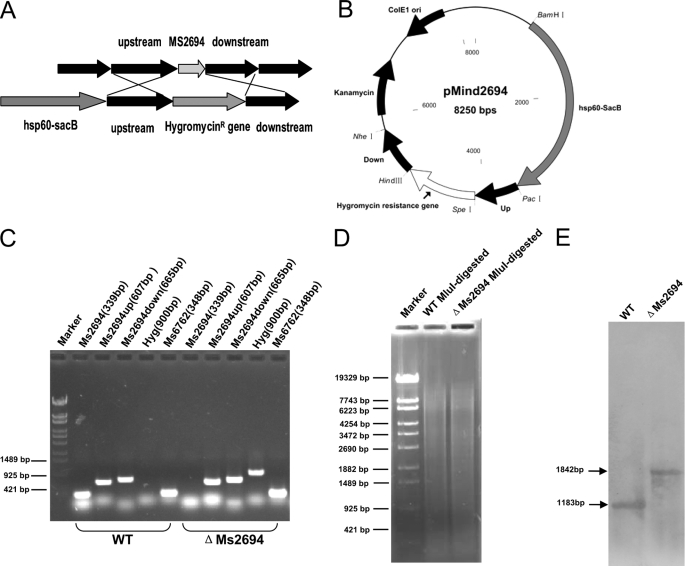
We produced a Ms2694-deletion mutant strain of M. smegmatis by a gene replacement strategy (Fig. 3, A and B) to further examine the regulation of DNA damage inducible genes by Ms 2694. A knock-out plasmid containing the up and down regions of the Ms2694 gene, and the selective hygromycin resistance gene (Hgr) was constructed and transformed into M. smegmatis. A ΔMs2694 strain, in which the Ms2694 gene was deleted (as confirmed by PCR results) was successfully produced by this method (Fig. 3C). A 900-bp Hgr gene fragment could be amplified by PCR from the genomic DNA of the ΔMs2694 strain but not from that of wild type M. smegmatis strain. In contrast, a 339-bp Ms2694 gene fragment could be amplified from the genomic DNA of wild type M. smegmatis but not from the ΔMs2694 strain. A Southern blot assay confirmed this result. As shown in Fig. 3, D and E, a signal band of ∼1.8 kb was detected (Fig. 3E) using a 317-bp probe from the MluI-digested genomic DNA of the mutant M. smegmatis strain. In contrast, a signal band of only ∼1.2 kb was seen in the wild type strain (Fig. 3E). This is consistent with the band sizes expected upon replacement of the Ms2694 gene with the Hygromycinr gene and indicated that the Ms2694 gene was successfully deleted in the mutant strain.
FIGURE 3.
Construction of the Ms2694 knock-out strain of M. smegmatis and Southern blot assays. A, schematic representation of the recombination strategy for removing Ms2694 from the genome of M. smegmatis. B, a map of the recombinant vector pMind2694 containing upstream and downstream sequences of Ms2694 and the gene that confers resistance against hygromycin. C, validation of the Ms2694 knock-out M. smegmatis strain. D, agarose gel electrophoresis of MluI-digested genomic DNA products from the Ms2694 knock-out M. smegmatis strain and the wild type strain. E, Southern blot assays. A 317-bp probe corresponding to the sequences of Ms2694 upstream genomic fragment of M. smegmatis was obtained by PCR and was labeled with digoxigenin dUTP (Roche Applied Science). The probe was used to detect the size change of the MluI-digested genomic fragment of M. smegmatis before and after recombination.
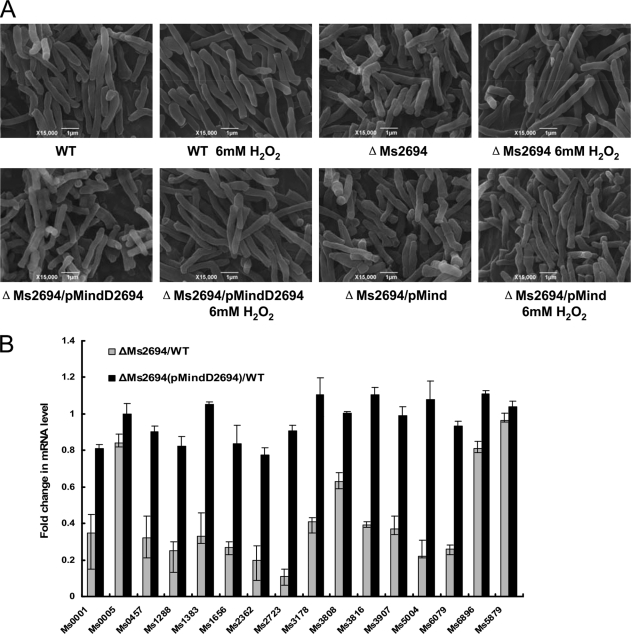
We used SEM to compare the morphological features of wild type and ΔMs2694 M. smegmatis. Ms2694 deletion did appear to alter the surface characteristics and the mutant cells were rough and irregular (Fig. 4A and supplemental Fig. S1). In contrast, wild type M. smegmatis cells had a smooth appearance. Another distinguishing morphological feature for the mutant M. smegmatis strain in comparison to wild type strain was increased cell lengths when induced by 6 mm H2O2 (Fig. 4A). We calculated lengths of the bacterial cells from representative fields as visualized by SEM (Table 1). Wild type M. smegmatis cells had lengths of 4.0 ± 1.5 μm and deletion of Ms2694 resulted in increased cell lengths of 6.0 ± 1.5 μm when induced by 6 mm H2O2 (Table 1). The p values of the cell length data were calculated and were <0.01. These differences had a statistical significance (Table 1). Transfection of mutant M. smegmatis with a pMind plasmid containing the Ms2694 gene (ΔMs2694/pMindD2694) restored the wild type growth pattern and morphology to the mutant strain, indicating that the phenotypic differences between ΔMs2694 and the wild type strain were due to loss of Ms2694. In contrast, the empty pMind plasmid without the Ms2694 gene did not rescue the aberrant ΔMs2694 phenotype (Fig. 4A).
FIGURE 4.
Cell growth and gene expression assays in the Ms2694-deficient M. smegmatis strain. A, bacterial morphology. Wild type and mutant M. smegmatis cells were prepared for SEM as described under “Experimental Procedures.” Morphological changes of M. smegmatis strains induced by 6 mm H2O2 were visualized by SEM, and images are representative of different fields of bacteria from exponentially growing cultures at 37 °C. Bars, 1 μm. B, quantitative real-time PCR assay for relative expression levels of DNA repair genes in the ΔMs2694 M. smegmatis strains. The myobacterial cDNA was amplified as described under “Experimental Procedures.” Relative expression levels of genes were normalized using sigA as an invariant transcript, and a non-RecA-NDp promoter gene Ms5879 was used as a negative control. Data were analyzed using the ΔΔCT method as described previously (23). As a positive control, the total DNA of each strain was used as the template for PCR amplification. The cDNA of a recombinant strain containing an empty pMind vector was used as a template in negative controls. p values of the relative expression data were calculated by unpaired two-tailed Student's t test using GraphPad Prism (version 5).
TABLE 1.
The cell length of wild type and mutant M. smegmatis strains with or without induction of 6 mm H2O2
The cells were examined by scanning electron microscope, and the sizes of 50 bacteria from each set were measured. The length measurement unit is micrometer (μm). The statistical significance (p values) of the cell length data was calculated by unpaired two-tailed Student's t test using GraphPad Prism (version 5). p values of the results (indicated by an asterisk) were <0.01.
| M. smegmatis mc2155 | M. smegmatis ΔMs2694 | M. smegmatis ΔMs2694 (pMindD2694) | M. smegmatis ΔMs2694 (pMind) | |
|---|---|---|---|---|
| No induction | 4.0 ± 1.0 | 3.5 ± 0.5 | 4.0 ± 0.5 | 3.5 ± 0.5 |
| H2O2 induction | 4.0 ± 1.5 | *6.5 ± 1.5 | 4.5 ± 1.0 | *6.0 ± 1.8 |
Relative gene expression levels in response to DNA damage were measured by quantitative RT-PCR. As shown in Fig. 4B, the expression of most of the tested genes containing the RecA-NDp motif was significantly down-regulated (p value < 0.01) after induction by 6 mm H2O2 for 3 h in the ΔMs2694 M. smegmatis strains compared with the wild type strain. In contrast, the expression of the negative control gene Ms5879 was not significantly different between the two strains. Strikingly, it was shown that we were able to complement the down-regulation of RecA-NDp genes in strain ΔMs2694/pMindD2694 (Fig. 4B) compared with the wild type strain. This indicated that Ms2694 could be a positive regulator or an inducer of genes regulated by promoters containing the RecA-NDp motif in response to DNA damage in M. smegmatis.
ClpR-like Factor of M. smegmatis Regulates Gene Mutation Frequencies and Rates
The ClpR-like factor of M. smegmatis, Ms2694, could directly bind to promoter regions of RecA-independent genes and positively regulate their expression in response to DNA damage. This strongly suggested that Ms2694 could regulate gene mutation frequencies in M. smegmatis. To examine this idea, we compared the gene mutation frequencies and rates in wild type and ΔMs2694 M. smegmatis strains. As shown in Table 2, the ΔMs2694 strains had a 7-fold higher streptomycin resistant gene mutation frequency (7.2 ± 1 × 10−9) compared with wild type M. smegmatis (1.2 ± 0.4 × 10−9). When expressing the Ms2694 gene through a pMind in ΔMs2694 strain, the recombinant strain of M. smegmatis ΔMs2694/pMindD2694 reobtained a similar resistant mutation frequency (0.8 ± 0.3 × 10−9) compared with the wild type strain (Table 2). The empty pMind plasmid had no effect on mutation frequency (Table 2). An assay of mutation rates using a fluctuation experiment obtained a similar result (Table 3, and supplemental Table S3). p values of the rates were calculated and only the value for the ΔMs2694 mutant was <0.05 if compared with wild type (Table 3). Thus, the difference was statistically significant. Ms2694 contributes significantly to genomic stability in M. smegmatis.
TABLE 2.
The mutation frequencies of wild-type and mutant M. smegmatis strains
| Strain | SM Mutation frequency×109 |
|---|---|
| M. smegmatis mc2155 | 1.2 ± 0.4 |
| M. smegmatis ΔMs2694 | 7.2 ± 1 |
| M. smegmatis ΔMs2694/pMindD2694 | 0.8 ± 0.3 |
| M. smegmatisΔMs2694/pMind | 8.0 ± 0.7 |
TABLE 3.
Mutation rates of M. smegmatis strains examined by fluctuation experiment
| Fluctuation experiment group number | Strain | No. of cultures | Nt valuea | Lea-Coulson m valueb | Mutation ratec |
|---|---|---|---|---|---|
| 1 | M. smegmatis mc2155 | 30 | 3.2×109 | 9 | 2.8×10−9 |
| 2 | M. smegmatis ΔMs2694 | 30 | 3.5×109 | 13 | *3.7×10−9 |
| 3 | M. smegmatis ΔMs2694/pMindD2694 | 30 | 4.5×109 | 11 | 2.4×10−9 |
a Final number of cells in the culture.
b Number of mutations per culture.
c Probability of mutation per cell per generation. p values of the results were calculated by unpaired two-tailed Student's t test using GraphPad Prism 5. p value of the result (indicated by an asterisk) was <0.05.
DISCUSSION
Like any organism, the intracellular pathogen M. tuberculosis possesses an inducible DNA repair system that responds to DNA damage, but unlike many other bacterial species, the identity of the regulators involved was unclear. In this study, we identified mycobacterial ClpR (Rv2745c in M. tuberculosis and Ms2694 in M. smegmatis mc2155) as a regulator that specifically binds to a conserved RecA-NDp motif both in vivo and in vitro. Compared with the wild type strain, the expression of these RecA-independent genes was down-regulated in the ClpR-deficient M. smegmatis strain in response to a DNA damage agent. These results provide important information for further understanding the function of the ClpR regulator and the regulatory mechanism of RecA/LexA-independent DNA repair in mycobacteria.
The RecA/LexA-dependent and RecA-independent pathways are the two known mechanisms of DNA repair in mycobacteria during the SOS response. In contrast to other species, the majority of inducible DNA repair genes in M. tuberculosis were found to RecA-independent (6–8). However, the molecular mechanisms of RecA-independent regulation of DNA repair genes had not yet been clearly defined in M. tuberculosis and related species. Using a bioinformatic approach, Gamulin et al. (12) mapped out a highly conserved RecA-NDp motif within the promoter regions of 47 RecA-independent genes in M. tuberculosis. However, a regulator that recognizes and binds to the conserved RecA-NDp motif within these promoter regions had not been characterized. In the current study, we conducted a bacterial one-hybrid assay in combination with EMSA assays and discovered that Rv2745c in M. tuberculosis and its M. smegmatis homolog Ms2694 could bind to the RecA-NDp motif. Furthermore, we demonstrated that the Ms2694 in M. smegmatis could bind to the promoter region of multiple DNA damage repair genes in vivo and thus regulate their expression in response to DNA damage. Our findings are consistent with several recent reports. For example, Rv2745c was shown to be induced in M. tuberculosis by multiple stress factors, including diamide stress (29), heat stress (30), SDS stress (31), hypoxia (32), infecting macrophages (31), vancomycin (33), and low pH (31). In addition, an anti-mycobacterial drug, thioridazine, which damages the bacterial cell wall, also induced Rv2745c expression (34). These reports indicate that Rv2745c plays an important role in the response to multiple environmental stress factors. Taken together with these results, the current observations strongly suggest that Rv2745c is a broad regulator of reparative genes that can be induced to activate the expression of DNA repair genes in response to stressful environmental signals and DNA damage agents in mycobacteria.
Rv2745c homologs have been found in all mycobacteria and in some other actinomycetes. They are usually called ClgRs and act as transcriptional activators of clp protease (13–15). An interesting finding of our work is that Ms2694, the M. smegmatis ClpR, regulates genomic stability by affecting gene mutation frequencies in M. smegmatis. This is consistent with a previous study in C. glutamicum in which the mRNA levels of the DNA repair genes recR, recN, and uvrA were found to decrease in a ClpR mutant strain compared with a wild type strain (16). Therefore, ClpR could be involved in expression of DNA repair genes in C. glutamicum although it was first characterized as a regulator of the clp protease gene (16). Interestingly, Rv2745c has also been found to play a role in regulating the clp genes in M. tuberculosis (17, 18). However, a recent study found that expression of the DNA repair genes recA and recG were induced, and two other DNA repair genes, uvrB and uvrD2, were repressed when Rv2745c was overexpressed in M. tuberculosis CDC1551 (31). Curiously, all four DNA repair genes contain the conserved RecA-NDp motif in their promoter sequences (12). Using the MycoRegNet web tool, which is based on the the C. glutamicum transcriptional network CoryneRegNet (35) for analyzing mycobacterial transcriptional regulatory networks, Krawczyk et al. (36) suggested that Rv2745c is involved in the SOS stress response and controls the expression of recR in M. tuberculosis. In addition, the recA gene in M. tuberculosis can be expressed through two different promoters, P1 and P2, both of which are DNA damage-inducible (9). Interestingly, only the P1 promoter contains the RecA-NDp motif (8, 9). In the present study, we provided evidence that Rv2745c and its homolog Ms2694 specifically recognized RecA-independent target genes, including RecA, in mycobacteria. Furthermore, by creating a ClpR-knock-out mutant strain, we found that the mycobacterial ClpR could respond to DNA damage and directly regulate expression of multiple DNA repair genes. These results, together with previous findings, strongly support the ClpR-like regulator as a core transcriptional factor that significantly contributes to damage-induced DNA repair in mycobacteria. These findings provide important insights into the function of ClpR regulators and the regulatory mechanism of RecA/LexA-independent DNA repair and gene mutation in mycobacteria.
Supplementary Material
Footnotes
This work was supported by the National Natural Science Foundation of China (Grants 30930003 and 31025002) and the Hubei Chutian Scholar Program (to Z.-G. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Fig. S1.
REFERENCES
- 1. World Health Organization (2010) Global Tuberculosis Control Report 2010 [Google Scholar]
- 2. Adams L. B., Dinauer M. C., Morgenstern D. E., Krahenbuhl J. L. (1997) Tuber. Lung Dis. 78, 237–246 [DOI] [PubMed] [Google Scholar]
- 3. Ehrt S., Schnappinger D. (2009) Cell Microbiol. 11, 1170–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Little J. W., Mount D. W. (1982) Cell 29, 11–22 [DOI] [PubMed] [Google Scholar]
- 5. Little J. W. (1982) Biochimie 64, 585–589 [DOI] [PubMed] [Google Scholar]
- 6. Rand L., Hinds J., Springer B., Sander P., Buxton R. S., Davis E. O. (2003) Mol. Microbiol. 50, 1031–1042 [DOI] [PubMed] [Google Scholar]
- 7. Brooks P. C., Dawson L. F., Rand L., Davis E. O. (2006) J. Bacteriol. 188, 6034–6038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dawson L. F., Dillury J., Davis E. O. (2010) J. Bacteriol. 192, 599–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davis E. O., Springer B., Gopaul K. K., Papavinasasundaram K. G., Sander P., Böttger E. C. (2002) Mol. Microbiol. 46, 791–800 [DOI] [PubMed] [Google Scholar]
- 10. Rauch P. J., Palmen R., Burds A. A., Gregg-Jolly L. A., van der Zee JR., Hellingwerf K. J. (1996) Microbiology 142, 1025–1032 [DOI] [PubMed] [Google Scholar]
- 11. Brooks P. C., Movahedzadeh F., Davis E. O. (2001) J. Bacteriol. 183, 4459–4467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gamulin V., Cetkovic H., Ahel I. (2004) FEMS Microbiol. Lett. 238, 57–63 [DOI] [PubMed] [Google Scholar]
- 13. Bellier A., Mazodier P. (2004) J. Bacteriol. 186, 3238–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Engels S., Schweitzer J. E., Ludwig C., Bott M., Schaffer S. (2004) Mol. Microbiol. 52, 285–302 [DOI] [PubMed] [Google Scholar]
- 15. Ventura M., Zhang Z., Cronin M., Canchaya C., Kenny J. G., Fitzgerald G. F., van Sinderen D. (2005) J. Bacteriol. 187, 8411–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Engels S., Ludwig C., Schweitzer J. E., Mack C., Bott M., Schaffer S. (2005) Mol. Microbiol. 57, 576–591 [DOI] [PubMed] [Google Scholar]
- 17. Sherrid A. M., Rustad T. R., Cangelosi G. A., Sherman D. R. (2010) PLoS One 5, e11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Estorninho M., Smith H., Thole J., Harders-Westerveen J., Kierzek A., Butler R. E., Neyrolles O., Stewart G. R. (2010) Microbiology 156, 3445–3455 [DOI] [PubMed] [Google Scholar]
- 19. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 20. Timm J., Post F. A., Bekker L. G., Walther G. B., Wainwright H. C., Manganelli R., Chan W. T., Tsenova L., Gold B., Smith I., Kaplan G., McKinney J. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14321–14326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guo M., Feng H., Zhang J., Wang W., Wang Y., Li Y., Gao C., Chen H., Feng Y., He Z. G. (2009) Genome Res. 19, 1301–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang M., Gao C., Wang Y., Zhang H., He Z. G. (2010) PLoS One 5, e10672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 24. Boshoff H. I., Reed M. B., Barry C. E., 3rd., Mizrahi V. (2003) Cell 113, 183–93 [DOI] [PubMed] [Google Scholar]
- 25. Rosche W. A., Foster P. L. (2000) Methods 20, 4–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Machowski E. E., Barichievy S., Springer B., Durbach S. I., Mizrahi V. (2007) J. Bacteriol. 189, 2190–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lea D. E., Coulson C. A. (1949) J. Genet. 49, 264–285 [DOI] [PubMed] [Google Scholar]
- 28. Hall B. M., Ma C. X., Liang P., Singh K. K. (2009) Bioinformatics 25, 1564–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mehra S., Kaushal D. (2009) J. Bacteriol. 191, 3965–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fontán P. A., Aris V., Alvarez M. E., Ghanny S., Cheng J., Soteropoulos P., Trevani A., Pine R., Smith I. (2008) J. Infect. Dis. 198, 877–885 [DOI] [PubMed] [Google Scholar]
- 31. Mehra S., Dutta N. K., Mollenkopf H. J., Kaushal D. (2010) J. Infect. Dis. 202, 943–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rustad T. R., Harrell M. I., Liao R., Sherman D. R. (2008) PLoS One 3, e1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Provvedi R., Boldrin F., Falciani F., Palù G., Manganelli R. (2009) Microbiology 155, 1093–1102 [DOI] [PubMed] [Google Scholar]
- 34. Dutta N. K., Mehra S., Kaushal D. (2010) PLoS One 5, e10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baumbach J., Brinkrolf K., Czaja L. F., Rahmann S., Tauch A. (2006) BMC Genomics 7, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krawczyk J., Kohl T. A., Goesmann A., Kalinowski J., Baumbach J. (2009) Nucleic Acids Res. 37, e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.