Abstract
Mutations in fibrillin-1 give rise to Marfan syndrome (MFS) characterized by vascular, skeletal, and ocular abnormalities. Fibrillins form the backbone of extracellular matrix microfibrils in tissues including blood vessels, bone, and skin. They are crucial for regulating elastic fiber biogenesis and growth factor bioavailability. To compare the molecular consequences of mutations causing the severe neonatal MFS with mutations causing the milder classical MFS, we introduced representative point mutations from each group in a recombinant human fibrillin-1 fragment. Structural effects were analyzed by circular dichroism spectroscopy and analytical gel filtration chromatography. Proteolytic susceptibility was probed with non-physiological and physiological proteases, including plasmin, thrombin, matrix metalloproteinases, and cathepsins. All mutant proteins showed a similar gross secondary structure and no differences in heat stability as compared with the wild-type protein. Proteins harboring neonatal mutations were typically more susceptible to proteolytic cleavage compared with those with classical mutations and the wild-type protein. Proteolytic neo-cleavage sites were found both in close proximity and distant to the mutations, indicating small but significant structural changes exposing cryptic cleavage sites. We also report for the first time that cathepsin K and V cleave non-mutated fibrillin-1 at several domain boundaries. Compared with the classical mutations and the wild type, the group of neonatal mutations more severely affected the ability of fibrillin-1 to interact with heparin/heparan sulfate, which plays a role in microfibril assembly. These results suggest differential molecular pathogenetic concepts for neonatal and classical MFS including enhanced proteolytic susceptibility for physiologically relevant enzymes and loss of function for heparin binding.
Keywords: Connective Tissue, Extracellular Matrix, Extracellular Matrix Proteins, Fibrillin, Heparin-binding Protein, Marfan Syndrome, Microfibrils, Mutations, Proteolysis
Introduction
Mutations in fibrillin-1 give rise to a number of heritable connective tissue disorders, termed fibrillinopathies. They include Marfan syndrome (MFS),3 stiff skin syndrome, autosomal dominant Weill-Marchesani syndrome, familial ectopia lentis, familial aortic aneurysm/dissection, and others (1, 2). The three human fibrillin genes code for large (350 kDa), extracellular, cysteine-rich proteins with a characteristic domain organization (3–5). The most prominent domains in fibrillins are the calcium binding epidermal growth factor like (cbEGF) domains and the 8-cysteine/transforming growth factor-β (TGF-β)-binding protein-like (TB) domains. These domains are stabilized by three or four intra-domain disulfide bonds, respectively (6–8). Fibrillins form the backbone of microfibrils in the extracellular matrix. Microfibrils are abundant in elastic and non-elastic connective tissues throughout the body, predominantly in blood vessels, lungs, skeletal elements, and skin. They confer structural integrity to individual organ systems, provide a scaffold for the biogenesis of elastic fibers, and regulate TGF-β signaling (3, 9).
The most common fibrillinopathy is MFS (Online Mendelian Inheritance in Man ID 154700), an autosomal dominant disorder with an estimated prevalence of 2–3 per 10,000 newborns (10). Major clinical symptoms are found in the cardiovascular (aortic aneurysm with dissection), ocular (ectopia lentis), and skeletal systems (long bone overgrowth, scoliosis). There are more than 1500 mutations described that are located throughout the entire fibrillin-1 gene (11). The most frequent type of mutations delete or introduce cysteine residues in various domains leading to at least one unpaired cysteine residue and an alteration of the disulfide bond pattern (1, 12). Another large class of point mutations affects the highly conserved calcium binding consensus sequence in cbEGF domains and interferes with calcium binding, typically rendering the molecule susceptible to proteolytic degradation (13–16). Inter- and intra-familial variability is a common feature of MFS, suggesting that other gene products play a modifying role in the individual etiopathology or that the consequences of a clinical mutation depend on the type of mutation and the context within the fibrillin-1 gene (1). Presently, no clear genotype-phenotype correlations have been established with the exception of the region covered by domains TB3-cbEGF18 (exons 24–32) in which mutations can cause early childhood mortality. This severe form of MFS is termed neonatal MFS (nMFS) (1). Classical MFS (cMFS) typically results from mutations elsewhere in the fibrillin-1 gene, although certain mutations located in the neonatal region have been shown to cause cMFS (17). Disease progression in MFS is the combined result of a loss in tissue integrity and perturbed TGF-β signaling (18–20). Loss of tissue integrity can result from deficiencies in the biogenesis of the microfibril/elastic fiber system and/or from excessive degradation of this system. Both mechanisms have been demonstrated with cells and tissues derived from patients with MFS (21–29).
There is growing evidence that proteases play a significant role in the pathophysiology of MFS. Calcium binding to cbEGF domains protects fibrillin-1 from proteolytic degradation by several physiological and non-physiological proteases in vitro (13, 14, 16, 30). It is documented that fibrillins are cleaved, for example, by matrix metalloproteinases (MMP)-2, -9, and -12 (31, 32). Some of the cleavage sites have been localized to exon 10/11 (proline-rich region-EGF4) and to the C-terminal end of cbEGF31 (31). None of these sites is located within the neonatal region of fibrillin-1. In other studies, the cMFS mutant E2447K but not E2169K, engineered in a fibrillin-1 C-terminal fragment, altered the degradation patterns for proteolysis with MMP-2, -9, -12, and -13 (32). This suggests the exposure of cryptic cleavage sites in mutant fibrillin-1 is dependent on the position of the mutation and not necessarily on the type of the amino acid exchanged. At the tissue level, MMP-1, -2, -3, and -9 proteases were up-regulated in thoracic aortas from MFS patients, and MMP-2 and -9 were up-regulated in a mouse model for MFS (33, 34). The addition of fibrillin-1 fragments containing the RGD integrin binding site to human dermal fibroblasts in cell culture induced the expression of MMP-1 and -3 (35). This suggests a potential vicious cycle where degraded fibrillin fragments may in turn lead to the further production of relevant proteases in tissues. Immunohistological studies showed a unique MMP and tissue inhibitor of metalloproteinase (TIMP) profile that emerged in ascending aortic aneurysms of individuals with MFS (33, 36). For example, MMP-12, membrane type-1 MMP, and TIMP-2 were up-regulated in these aortic tissues. Cathepsins, known for their intracellular function in lysosomes, have also been detected in the extracellular environment. Cathepsin K and V have elastolytic activity and have been identified in macrophages present in plaques of diseased blood vessels, in osteoclasts facilitating bone resorption, and in the progression of artheriosclerotic plaques in cerebral blood vessels (37–39). Nothing is known about proteolytic activity of cathepsin K and cathepsin V on fibrillin molecules. In summary, the published evidence suggests that entire or partial proteolytic degradation of fibrillin and microfibrils are involved in loss of tissue integrity. This may consequently lead to fewer functional microfibrils in tissues associated with elevated levels of active TGF-β, which significantly contributes to the pathogenesis of MFS (3, 18, 40).
The aim of the current study is to analyze functional consequences of mutations in fibrillin-1 that cause nMFS and to compare them to mutations that cause cMFS. We expressed and purified several nMFS and cMFS mutations within a centrally located fragment of fibrillin-1 harboring the neonatal region and analyzed the susceptibility for proteolytic degradation using a panel of non-physiological and physiological proteases. In addition, we analyzed the functional consequences of the mutations on heparin binding. We found that generally the nMFS mutations had more severe consequences on proteolytic susceptibility as well as on the interaction with heparin. The results from this study may suggest novel pathogenetic players contributing to MFS and other fibrillinopathies.
EXPERIMENTAL PROCEDURES
Cloning Strategies and Site-directed Mutagenesis
The rF20 fragment used in this study includes a sequence coding for an N-terminal BM-40 signal peptide to ensure proper secretion in human embryonic kidney cells (HEK 293) and a C-terminal hexahistidine tag to facilitate purification. The cloning and production of the rFBN1-N and rFBN1-C fragment were described previously (41, 42). All of the mutations analyzed in this study were reported in patients with MFS and are listed in the Universal Mutation Database (11). Generation of the mutation plasmids relied on a number of previously published plasmids mentioned below (13, 30, 43). The plasmid coding for Asp451–Val1527 of fibrillin-1 was generated by restricting pcDFRSP-rF45-WT and pCEPSP-rF18 with Bsu36I × NotI of which the 6629- and the 1727-bp fragment, respectively, were ligated to generate the plasmid coding for the rF20-WT construct (pDNSP-rF20-WT). The same strategies were used to generate plasmids coding for rF20-C750G (pcDFRSP-rF45-C750G plus pCEPSP-rF18) and rF20-R627C (pcDFRSP-rF45-R627C plus pCEPSP-rF18). The 7185-bp NheI × Bsu36I fragment from pDNSP-rF20-WT was ligated with the 1534-bp NheI × Bsu36I fragment from pCEPSP-rF45-N548I, resulting in the plasmid coding for rF20-N548I. To generate mutant polypeptide rF20-E1073K, a 6992 bp Bsu36I × NotI fragment from pDNSP-rF20-WT was ligated with a 1727 bp Bsu36I × NotI fragment from pCEPSP-rF18-E1073K. Mutation constructs rF20-G1013R, rF20-C1032Y, rF20-I1048T, and rF20-C1182S were generated using the QuikChange site-directed mutagenesis kit (Stratagene) with pDNSP-rF20-WT as a template according to the supplied protocol. The fibrillin-1 oligonucleotide primers harboring the single nucleotide exchanges are listed in supplemental Table 1. The presence of the mutations in all constructs was confirmed by DNA sequencing.
Cell Culture and Transfection
HEK 293 were grown in DMEM supplemented with 10% fetal calf serum, 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin in a humidified incubator at 37 °C in a 5% CO2 atmosphere. 10 μg of the linearized plasmid DNA of the different rF20 constructs was transfected into HEK 293 cells with Lipofectamine 2000 according to the supplied protocol (Invitrogen). Two days after transfection, the selection procedure was initiated by supplementing the medium with 0.25 mg/ml G418 sulfate (Wisent). Secretion of the rF20 proteins was analyzed by SDS-PAGE and Western blotting after trichloroacetic acid precipitation. Conditioned serum-free medium with secreted rF20 mutant proteins was collected from triple layer flasks (Nunc) and supplemented with protease inhibitors (0.1 mm PMSF, 1 tablet/500 ml of EDTA-free Complete, Roche Applied Science).
Purification of Recombinant Proteins
The proteins were purified to homogeneity using an ÄktaPurifier (GE Healthcare) with the following purification schemes; the pH of 2 liters of conditioned medium was adjusted to 8.8 with 2 m Tris-base and filtered through a 5-μm membrane filter (Millipore). A 50-ml Q-Sepharose HP anion exchange column (GE Healthcare) was equilibrated with 50 mm Tris-HCl, pH 8.8, and the medium was applied at a flow rate of 10 ml/min. Bound protein was eluted with 20 mm HEPES, 1 m NaCl, pH 7.2, directly onto a 5-ml HisTrap HP column (GE Healthcare) loaded with Ni2+ and equilibrated in the same buffer. Bound proteins were eluted from the HisTrap column with a gradient of 0–500 mm imidazole in 20 mm HEPES, 1 m NaCl, pH 7.2. Fractions were analyzed by SDS-PAGE, and the ones containing the purified protein were pooled, treated with 5 mm EDTA, and dialyzed against TBS (50 mm Tris-HCl, 150 mm NaCl, pH 7.4) including 2 mm CaCl2 (TBS-Ca) and stored at −80 °C. Protein concentrations were determined using the BCA Protein Assay kit (Pierce). To improve the purity, an additional preparative gel filtration chromatography step was occasionally required for the mutant polypeptides rF20-N548I and rF20-C1032Y. A 110-ml Superose-12 gel filtration column (GE Healthcare) was equilibrated with 400 mm NaCl, 50 mm Tris-HCl, 5 mm CaCl2, pH 7.4, at 0.5 ml/min. Samples were concentrated to ∼1.5 ml using a Centriplus YM-30 unit (Millipore), loaded onto the column at a flow rate of 0.5 ml/min, and fractionated (1 ml fractions). Fractions containing the purified proteins were identified via SDS-PAGE, pooled, and dialyzed against TBS-Ca. Western blotting with specific mono- and polyclonal antibodies as well as N-terminal sequencing using Edman degradation were used to confirm the identity and integrity of purified rF20 proteins.
Gel Filtration Chromatography
A analytical Superose 12 300GL column (GE Healthcare) was equilibrated in 50 mm Tris-HCl, 150 mm NaCl, 2 mm CaCl2, pH 7.4, and operated with an ÄktaPurifier (GE Healthcare). 200 μg of protein was loaded onto the column at a flow rate of 1 ml/min, and 0.5-ml fractions were collected. 20-μl aliquots of selected peak fractions were subjected to SDS-PAGE under non-reducing and reducing (0.1 m dithiothreitol, DTT) conditions and visualized by silver staining.
Circular Dichroism Spectroscopy
Circular dichroism (CD) spectroscopy was used to analyze global alterations in protein secondary structure. Measurements were obtained in a 1 mm quartz cuvette using a Jasco J-815 CD spectropolarimeter. Samples were dialyzed against 1 mm KH2PO4, 6.6 mm NaH2PO4, 0.6 mm CaCl2, pH 7.0, and CD spectra were recorded using a protein concentration of 0.3 mg/ml (13, 44). Because an accurate protein concentration is critical for the correct calculation of the molar ellipticity, the protein concentrations were determined directly before CD measurements using the BCA assay. The spectrum of each polypeptide was measured at 20 and 95 °C with a scan speed of 50 nm/min. An average spectrum was obtained from five repeats. The percentage of secondary structure elements was calculated after deconvolution of the spectra using the Contin LL algorithm from the DichroWeb server (45, 46). Thermal denaturation profiles were measured with a heating rate of 0.4 °C/min at 204 nm, where the greatest difference in the mean molar ellipticity between 20 and 95 °C was observed. Measurements were converted to percent denatured protein, where the ellipticity at 20 °C was set to 0% denatured and the ellipticity at 95 °C was set to 100%.
Proteolytic Assays
For proteolytic degradation assays, 7 μg of each rF20 polypeptide was digested with non-physiological (trypsin and chymotrypsin) and physiological (plasmin, thrombin, cathepsin K and V, and MMP-1, -2, -3, -9, and -12) proteases at various enzyme to protein ratios for up to 24 h at 37 °C. Degradation patterns were analyzed by reducing SDS-PAGE using 4–20% gradient gels. Optimized enzyme-to-protein ratios and incubation times were used as follows: trypsin and chymotrypsin = 1:100 (w/w) for 30 min, plasmin and thrombin = 1:20 (w/w) for 24 h, cathepsin K = 1:50 (w/w) for 6 h, cathepsin V = 1:100 (w/w) for 6 h, and MMPs = 1:100 (w/w) for 24 h. Trypsin, chymotrypsin, plasmin, and thrombin were purchased from Sigma, MMP-1,-2,-3,-9, and -12 were obtained from R&D systems. MMPs were activated using 4-aminophenylmercuric acetate (Sigma) or by proteolytic self-activation according to the manufacture's specifications (Sigma). Cathepsin K and V were expressed in a non-glycosylated form in the yeast Pichia pastoris and activated with 0.1 m sodium acetate at pH 5.5 as described previously (47, 48). All enzymes were stored at −80 °C in aliquots to avoid freeze-thaw cycles. After proteolysis, the enzymes were deactivated by heat denaturation at 95 °C and 20 mm DTT, and the fragments were separated on 4–20% gradient SDS-PAGE followed by Coomassie Brilliant Blue staining. To characterize the degree of fragmentation, the intensity profile of individual lanes of scanned gels was measured using the Molecular Imaging Software Version 4.0 (Eastman Kodak Co.). The background corrected net intensity profiles were plotted as three-dimensional graphs using the Origin 7.0 software package (OriginLab).
N-terminal Sequencing
Protein samples (7.2–64 μg) were incubated with the respective enzymes, separated by SDS-PAGE, and transferred onto a polyvinylidene fluoride membrane (Bio-Rad) using 10 mm sodium borate at 0.4 A for 1.5 h. Blots were stained with Coomassie Brilliant Blue in 50% methanol and 5% acetic acid for 5 min and destained with 50% methanol and 5% acetic acid for 2 × 5 min. The blot was thoroughly washed with distilled water and air-dried. Bands were excised and subjected to N-terminal sequencing by Edman degradation using an automated peptide sequencer (Model Procise 494, Applied Biosystems). Sequences were analyzed using the SequencePro 2.0 software (Applied Biosystems). Bands, of which the N-terminal sequence could be identified, are labeled with numbers that correlate to the numbers in Tables 3 and 4.
TABLE 3.
Neo-cleavage sites in rF20 mutant proteins identified by N-terminal sequencing
For numbering of the amino acids, fibrillin-1 sequence NP_000129.3 was used. The number for the position in FBN1 refers to the first residue after the cleavage site (P1′). Eight residues were identified in each N-terminal sequencing reaction, which is sufficient to unambiguously identify the cleavage site (P1′–P8′). Cysteine can normally not be analyzed by Edman degradation. However, frequently the stable cysteine-acryl amid compound could be identified in the chromatograms (67). Degradation sites also found in the WT were omitted. For trypsin and chymotrypsin, only cleavages sites corresponding to the consensus sites are indicated. The region from residue 1013 to 1027 represents a relatively long linker between TB3 and cbEGF11. T, trypsin; C, chymotrypsin; P, plasmin; Th, thrombin; MMP, matrix metalloproteinase; Cat, cathepsin.
| Mutation | Cleavage site |
Band number | Position in FBN1 | Position relative to mutation | Module | Enzyme | |
|---|---|---|---|---|---|---|---|
| P4 - P1 | P1′ - P8′ | ||||||
| N548I | PGLA | VGLDGRVC | 5 | 645 | +97 | cbEGF6 | Cat K |
| R627C | LAVG | LDGRVCVD | 6 | 647 | +20 | cbEGF6 | P |
| GKNC | VDINECVL | 11 | 764 | +137 | cbEGF7 | Th | |
| C750G | ENLR | GTYKCIGN | 8 | 744 | −6 | cbEGF7 | Th |
| RGTY | KCIGNSGY | 9 | 747 | −3 | cbEGF7 | C | |
| GTYK | CIGNSGYE | 10 | 748 | −2 | cbEGF7 | P | |
| CVLN | SLLCDNGQ | 12 | 773 | +23 | cbEGF8 | MMP-3 | |
| GQCR | NTPGSFVC | 13 | 783 | +33 | cbEGF8 | T | |
| GMTL | DATGRICL | 14 | 944 | +194 | cbEGF10 | C | |
| DMGY | SGKKTKGC | 25 | 1312 | +562 | cbEGF17 | C | |
| G1013R | QLVR | YLCQNGRC | 1 | 458 | −555 | EGF4 | T |
| QSTL | TRTECRDI | 2 | 524 | −489 | cbEGF3 | C | |
| TLTR | TECRDIDE | 3 | 526 | −487 | cbEGF3 | T | |
| RPGP | FATKEITN | 16 | 1014 | +1 | TB3 | Th | |
| PGPF | ATKEITNG | 17 | 1015 | +2 | TB3 | MMP-12 | |
| ATKE | ITNGKPFF | 18 | 1019 | +6 | TB3 | MMP-12 | |
| GKPF | FKDINECK | 19 | 1026 | +13 | TB3 | MMP-3 | |
| PFFK | DINECKMI | 21 | 1028 | +15 | TB3 | T | |
| C1032Y | QLVR | YLCQNGRC | 1 | 458 | −574 | EGF4 | T |
| DSGF | ALDSEERN | 22 | 1060 | +28 | cbEGF11 | C | |
| I1048T | QLVR | YLCQNGRC | 1 | 458 | −590 | EGF4 | T |
| KPFF | KDINECKM | 20 | 1027 | −21 | TB3 | Cat V | |
| DMGY | SGKKTKGC | 25 | 1312 | +246 | cbEGF17 | C | |
| E1073K | QLVR | YLCQNGRC | 1 | 458 | −615 | EGF4 | T |
| QSTL | TRTECRDI | 2 | 524 | −549 | cbEGF3 | C | |
| TLTR | TECRDIDE | 3 | 526 | −547 | cbEGF3 | T | |
| DSGF | ALDSEERN | 22 | 1060 | −13 | cbEGF11 | C | |
| DECR | ISPDLCGR | 23 | 1076 | +3 | cbEGF12 | T | |
| KKGK | TGCTDINE | 26 | 1318 | +245 | cbEGF17 | T | |
| C1182S | QLVR | YLCQNGRC | 1 | 458 | −724 | EGF4 | T |
| RGPG | FATKEITN | 16 | 1014 | −168 | TB3 | Cat K | |
| LIGK | YQCACNPG | 24 | 1178 | −4 | cbEGF14 | MMP-3 | |
TABLE 4.
Cathepsin K and V cleavage sites in non-mutated recombinant fibrillin-1 fragments identified by N-terminal sequencing
The details for the fibrillin-1 sequence and numbering are identical to the legend for Table III.
| Fragment | Cleavage site |
Band number | Position in FBN1 | Domain | |
|---|---|---|---|---|---|
| P4–P1 | P1′–P12′ | ||||
| Cathepsin K | |||||
| rFBN1-N | CGSR | SIQHCNIRCMNG | 6 | 115 | Linker EGF1-EGF2 |
| EPPR | VLPVNVTDYCQL | 14 | 444 | End of proline-rich | |
| PPRV | LPVNVTD | 12 | 445 | End of proline-rich | |
| PGMT | SAGSDINECALD | 4 | 719 | Linker TB2-cbEGF7 | |
| rFBN1-C | GNCT | DVNECLDP | 5 | 1487 | N-terminus of fragment |
| Cathepsin V | |||||
| rFBN1-N | GSRS | IQHCNIRCMNGG | 7 | 116 | Linker EGF1-EGF2 |
| PGMT | SAGSDINECALD | 2 | 719 | Linker TB2-cbEGF7 | |
| rFBN1-C | GNCT_ | DVNECLDP | 1 | 1487 | N-terminus of fragment |
| CPYG | SGIIVGPDDSAV | 8, 9, 10, 11 | 2115 | Linker TB6-cbEGF32 | |
Heparin Binding
A 1 ml HiTrap Heparin HP column was operated by an ÄktaPurifier (GE Healthcare) and equilibrated in 20 mm Tris, 50 mm NaCl, 2 mm CaCl2, pH 7.4 (49). 150 μg of protein in equilibration buffer was applied to the column at a flow rate of 0.1 ml/min. Bound protein was eluted with a gradient of 0–500 mm NaCl in the same buffer, and aliquots of the fractions were analyzed by SDS-PAGE under reducing and non-reducing conditions followed by silver staining. The protein bands in the flow-through and in the bound fraction were quantified by ImageJ version 1.44 (50).
RESULTS
Recombinant Mutant Fibrillin-1 Proteins
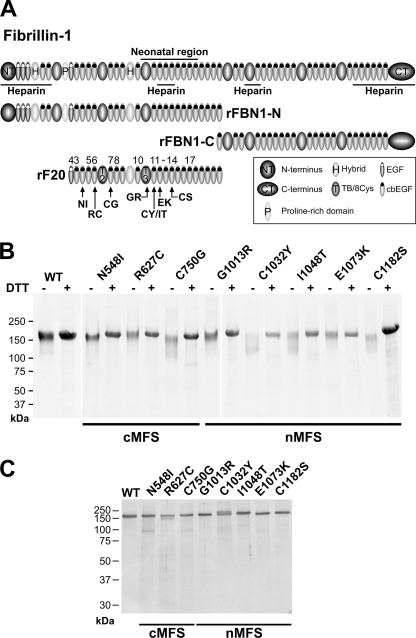
For the analysis of structural and functional consequences of mutations in fibrillin-1, we introduced a series of mutations causing cMFS (N548I, R627C, C750G) and nMFS (G1013R, C1032Y, I1048T, E1073K, C1182S) in the recombinant fibrillin-1 fragment rF20, which was previously shown to be effectively secreted from HEK 293 cells (Table 1, Fig. 1A) (43). This construct spans the region from EGF4 to cbEGF22 (amino acid position 451–1527) of human fibrillin-1 and can be readily purified by a C-terminal hexahistidine tag and Ni2+-chelating chromatography. This strategy enables for the analysis of the consequences of mutations in a larger and post-translational modified protein context compared with relatively small recombinant fibrillin-1 fragments used in other studies. We did not attempt to analyze the mutations in full-length fibrillin-1 because it is very difficult to produce and purify, and it has the propensity to aggregate (41). Mutations were selected to cover different principal categories, including mutations that affect calcium binding (N548I, E1073K), or interfere with the disulfide-bond patterning by introducing (R627C) or deleting (C1032Y, C750G, C1182S) cysteine residues. Mutations not involved in the former groups (G1013R, I1048T) were also included. The mutated fragments were secreted from the recombinant cell clones and purified to homogeneity (Fig. 1B).
TABLE 1.
Mutations in FBN1 analyzed in this study
| Amino acid exchange | Nucleotide exchange | Exon | Domain | Clinical consequence | Reference no. |
|---|---|---|---|---|---|
| N548I (NI) | 1643A→T | 13 | cbEGF4 | Classical | (62) |
| R627C (RC) | 1879C→T | 15 | cbEGF6 | Classical | (63) |
| C750G (CG) | 2248T→A | 18 | cbEGF7 | Classical | (63) |
| G1013R (GR) | 3037G→A | 24 | TB3 | Neonatal | (64) |
| C1032Y (CY) | 3095G→A | 25 | cbEGF11 | Neonatal | (65) |
| I1048T (IT) | 3143T→C | 25 | cbEGF11 | Neonatal | (65) |
| E1073K (EK) | 3217G→A | 26 | cbEGF12 | Neonatal | (64) |
| C1182S (CS) | 3545G→C | 28 | cbEGF14 | Neonatal | (66) |
FIGURE 1.
Fibrillin-1 protein fragments used in this study. A, domain organization of the fibrillin-1 protein and the rF20 fragment is shown. The neonatal region and the four heparin binding sites in fibrillin-1 are indicated. Individual domains are represented as ellipses and described. The position of each generated MFS point mutation within the rF20 polypeptide is indicated by arrows. Mutations N548I (NI), R627C (RC), C750G (CG) cause the cMFS, and mutations G1013R (GR), C1032Y (CY), I1048T (IT), E1073K (EK), C1182S (CS) cause the nMFS. B, shown is a demonstration of purity of the protein preparations. The mutant rF20 proteins were purified to homogeneity as described under “Experimental Procedures.” 7 μg per lane were analyzed by SDS-PAGE under reducing (DTT+) and non-reducing (DTT−) conditions and stained with Coomassie Brilliant Blue. C, to test for potential residual proteolytic activity in the purified proteins samples, the proteins were incubated for 24 h at 37 °C and analyzed as described in B under reducing conditions. Positions of globular marker proteins are indicated in kDa.
Structural Analysis of Mutant Fibrillin-1 Proteins
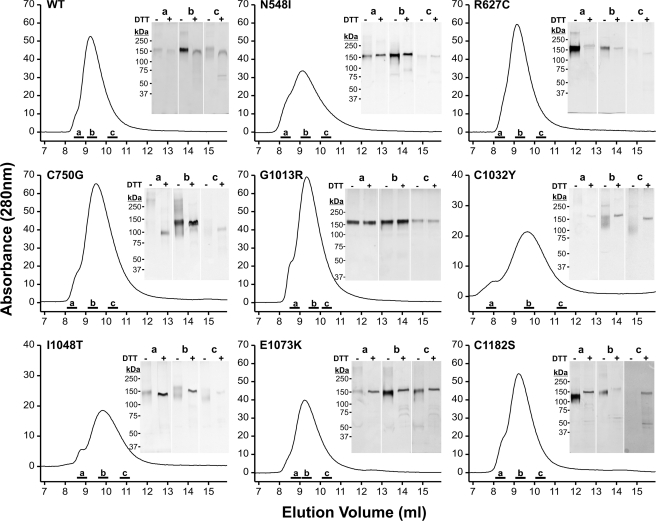
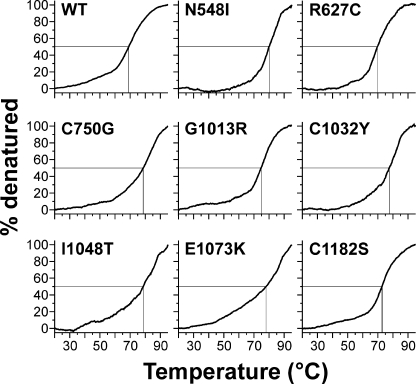
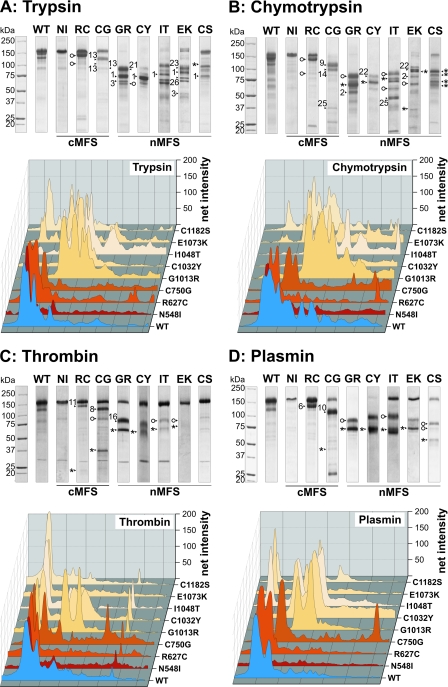
Analysis by SDS-PAGE under reducing conditions showed that all mutant proteins had identical electrophoretic properties compared with the wild-type (WT) protein (Fig. 1B). Similar migration for all mutant proteins was also observed under non-reducing conditions, with C750G, C1032Y, I1048T, and C1182S mutant proteins showing a slightly broader protein band. This may result from an altered glycosylation pattern or from a more compact structure. We further analyzed the status of the proteins with respect to dimerization or aggregation by analytical gel filtration chromatography (Fig. 2). All mutant proteins eluted primarily as monomers with very small amounts of reducible multimers. Correlating with the SDS-PAGE, the elution peaks for the C750G, I1048T, and C1032Y shifted slightly to higher elution volumes either from reduced glycosylation or from a somewhat more compact shape. CD spectroscopy was employed to examine overall secondary structural changes and destabilizing effects induced by the individual mutations. For all mutant proteins and the control protein, the minima of the molar ellipticities at 20 °C were identified between 208 and 211 nm (Table 2). These results are characteristic for fibrillin-1 harboring 43 cbEGF domains, which contain significant amounts of β-structures (13, 30). Although the absolute degrees of ellipticity varied at the respective minima, relatively little overall changes in the secondary structures were observed after deconvolution using the Contin L algorithm. As expected, β-strand conformations were most prevalent (29.3–34.6%), whereas α-helices (11.0–18.2%) and β-turns (12.4–13.2%) contributed less to the secondary structures (Table 2). In thermal denaturation experiments, the largest temperature-dependent differences in molar ellipticities were observed at 204 nm (data not shown). To determine whether the mutations destabilize the proteins, denaturation profiles from 20–95 °C were recorded at 204 nm (Fig. 3). The unmodified rF20-WT was denatured to 50% at ∼70 °C (Table 2). All mutant fibrillin-1 proteins displayed similar profiles where 50% denaturation was reached between 71.0 and 77.5 °C. These results show that the individual point mutations did not reduce the overall stability of rF20 and that the region in fibrillin-1 covered by rF20 is generally very stable against thermal denaturation. In summary, the analyses by CD spectroscopy demonstrate that the mutations do not cause extensive global changes in secondary structure. In addition, analyzing the splicing of X-box-binding protein-1 did not indicate induction of the unfolded protein response in the endoplasmic reticulum either for WT or for mutant proteins (data not shown) (51). Because the mutant proteins displayed no gross secondary structure alterations, we set out to probe for local structural changes by analyzing protease neo-cleavage sites, which are normally not accessible in the rF20-WT construct. For this purpose, we used the non-physiological proteases trypsin and chymotrypsin, which have many predicted cleavage sites in the primary rF20 sequence. On average, there is one trypsin cleavage site per 12.8 residues and one chymotrypsin cleavage site per 7.8 residues in rF20. To test for potential contamination of the purified proteins by proteases, samples were incubated at 37 °C for 24 h, which represents the longest incubation time used in the following protease degradation experiments (Fig. 1C). None of the mutant proteins displayed significant degradation. Treatment of the proteins harboring nMFS mutations with either trypsin or chymotrypsin consistently resulted in more fragmentation (>80%) compared with those with cMFS mutations or the rF20-WT (Fig. 4, A and B). Generally, the mutant proteins with nMFS mutations degraded more readily accompanied with the production of more neo-cleavage sites. For the mutations C750G, G1013R, C1032Y, and E1073K, trypsin and chymotrypsin neo-cleavage sites were identified by N-terminal sequencing directly in the mutated domain, illustrating that the introduced mutations modify the local structural properties (Table 3). In addition, neo-cleavage sites were also observed at a distance farther away from the mutated domain, indicating long range structural effects for all tested nMFS mutations and for the cMFS C750G mutation (Table 3). The trypsin site observed at amino acid position 458 is located in the N-terminal EGF module of the mutant rF20 proteins. This sensitive site likely arises from domain instability at the N terminus rather than from structural alterations by the respective mutations (52, 53). Generally, identical cleavage sites that were also found in the wild-type rF20 were not considered neo-cleavage sites for trypsin and chymotrypsin as well as for the physiological proteases described below. These sites are summarized in supplemental Table 2.
FIGURE 2.
Gel filtration chromatography of the mutant rF20 polypeptides. The rF20 polypeptides were subjected to gel filtration chromatography, as detailed under “Experimental Procedures,” to separate monomers from potentially present multimers. Elution profiles at A280 nm are shown. Aliquots of representative peak fractions labeled by bars (a, b, and c) were separated by SDS-PAGE under non-reducing (DTT−) and reducing (DTT+) conditions as indicated and silver-stained. Note that the silver-staining procedure stains reduced proteins to a lesser degree than non-reduced proteins. The positions of globular marker proteins are indicated in kDa. Each fraction was analyzed on the same gel under both conditions except for G1013R (b), for which samples were loaded on two gels.
TABLE 2.
Circular dichroism analysis of WT and mutant rF20 polypeptides
| Mutation | Wavelength minimum | Molar ellipticity at minimum | T at 50% denatured | Secondary structure elements. |
|||
|---|---|---|---|---|---|---|---|
| α-Helix | β-Strand | β-Turn | Unordered | ||||
| nm | degree × cm2 × dmol−1 | ° C | % | ||||
| WT | 209.9 | −5512.4 | 71.0 | 11.1 | 34.3 | 13.0 | 41.6 |
| N548I | 209.6 | −6901.9 | 77.2 | 18.2 | 29.3 | 12.9 | 39.6 |
| R627C | 211.0 | −5433.1 | 71.7 | 12.9 | 33.0 | 13.0 | 41.2 |
| C750G | 210.3 | −6431.2 | 77.5 | 15.5 | 30.9 | 13.1 | 40.6 |
| G1013R | 210.6 | −5411.3 | 73.1 | 11.0 | 34.3 | 13.0 | 41.7 |
| C1032Y | 208.1 | −5223.2 | 75.4 | 11.9 | 33.9 | 13.1 | 41.3 |
| I1048T | 210.7 | −5507.2 | 76.2 | 14.0 | 30.7 | 13.2 | 42.1 |
| E1073K | 210.2 | −5393.5 | 76.4 | 11.1 | 34.6 | 12.8 | 41.5 |
| C1182S | 209.5 | −5361.4 | 73.1 | 13.7 | 32.8 | 12.4 | 41.1 |
FIGURE 3.
Thermal denaturation of rF20 polypeptides assessed by circular dichroism spectroscopy. Denaturation profiles were recorded at 204 nm, and the proteins were considered as 100% denatured at 95 °C. For all proteins analyzed, 50% denaturation was observed at similar temperatures between 71 and 77.5 °C (crossing point of the horizontal and vertical lines), indicating that the mutations did not have a profound effect on the overall structural stability of the polypeptide.
FIGURE 4.
Proteolysis of rF20 polypeptides with various non-physiological and physiological proteases. 7 μg of each polypeptide was digested with trypsin (A), chymotrypsin (B), thrombin (C), and plasmin (D) as detailed under “Experimental Procedures.” Proteolytic degradation products were separated by SDS-PAGE under reducing conditions and stained with Coomassie Brilliant Blue (upper panels). The numbered arrows correlate with the determined N-terminal sequences of the degradation products summarized in Table 3. The abbreviations of mutations are used according to Table 1. Mutations leading to classical Marfan syndrome are indicated as cMFS, and mutations leading to neonatal Marfan syndrome are indicated as nMFS. Fragments starting with the “APLADYCQ” amino acid sequence (N terminus of rF20) are labeled with an asterisk (*). Cleavage sites marked with an open circle were also found in the wild-type rF20 (WT). The lanes for each of the degradation analyses were analyzed by densitometry and plotted as a three-dimensional graph (lower panels). The profile of the WT is depicted in blue, the cMFS mutations is in shades of red, and the nMFS mutations are in shades of beige. High molecular weight bands are positioned on the left.
Susceptibility of Mutant Proteins to Physiological Proteases: Thrombin and Plasmin
Thrombin and plasmin were used to probe for susceptible sites in the mutant proteins because these proteases are found in extracellular matrices where fibrillin-1 and microfibrils are expressed and whereby plasmin, but not thrombin, has been shown to cleave fibrillin-1 (13, 16). The proteins harboring nMFS mutations G1013R, C1032Y, and I1048T and the classical mutation C750G showed enhanced degradation by thrombin evidenced by either more or different neo-cleavage bands occurring after proteolytic degradation as compared with the WT (Fig. 4C). Other mutant proteins were only slightly affected if at all. After plasmin treatment, all mutant proteins except rF20-N548I demonstrated neo-cleavage products (Fig. 4D). Typical and almost complete degradation of the neonatal rF20-G1013R and rF20-C1032Y indicate that the respective mutations render the protein particularly sensitive to degradation with plasmin. For both thrombin and plasmin, the identified neo-cleavage sites that are not present in the non-mutated rF20-WT are located within the mutated domain or in adjacent domains (Table 3).
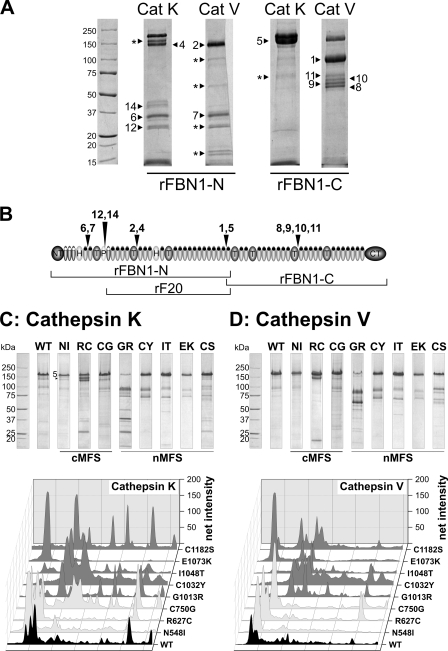
Cathepsin K and V
Cathepsins K and V have been located in extracellular matrices and are highly elastolytic (37, 54). However, it is not known whether these cathepsins have the potential to degrade fibrillin-1. Using recombinant cathepsins to degrade the non-mutated N-terminal (rFBN1-N) and C-terminal (rFBN1-C) halves of fibrillin-1, we found that cathepsin K readily cleaved rFBN1-N at multiple sites, whereas it cleaved rFBN1-C only at one major site resulting in a double band (Fig. 5, A and B, Table 4). Cathepsin V readily cleaved both rFBN1-N and rFBN1-C at multiple cleavage sites. Interestingly, all identified cleavage sites are located either in linker regions between two domains or at the end of the proline-rich domain such that the highly disulfide-bonded fibrillin-1 structure would come apart if cleavage occurs at these sites as opposed to intra-domain cleavages (Table 4). This is the first description of its kind, demonstrating that fibrillin-1 is a target for cathepsin K and V. Degradation of the mutant fibrillin-1 proteins with cathepsin K and V typically resulted in a different and enhanced fragmentation pattern for the constructs harboring nMFS mutations compared with the non-mutated rF20-WT and the mutant proteins harboring cMFS mutations (Fig. 5, C and D). rF20-G1013R was the most sensitive to proteolysis for both cathepsin K and V. The non-mutated and two of the three cMFS mutant proteins (N548I, C750G) were relatively resistant to degradation by both cathepsins. The R627C mutant protein was the only protein harboring a cMFS mutation that showed significant degradation. In an attempt to obtain sequence information, nine degradation products were identified as the N terminus of the respective proteins (data not shown). Other degraded protein bands were not interpretable.
FIGURE 5.
Cleavage of non-mutated and mutated fibrillin-1 fragments with cathepsin K and cathepsin V. A, the N- and C-terminal half of non-mutated fibrillin-1 (rFBN1-N, rFBN1-C, respectively) was cleaved with cathepsin K (Cat K) and cathepsin V (Cat V) as outlined under “Experimental Procedures.” The numbered arrows correlate with the determined N-terminal sequences of the degradation products summarized in Table 4. Fragments labeled with an asterisk (*) could not be identified by N-terminal sequencing. Bands 1 and 5 represent the N-terminal APLA sequence of the rFBN1-C fragment. B, shown is schematic mapping of the cathepsins K and V cleavage sites identified in A. The domains are annotated as in Fig. 1. C and D, wild-type and mutant rF20 was digested with cathepsins K and V as indicated. Proteolytic degradation products were separated by SDS-PAGE under reducing conditions and stained with Coomassie Brilliant Blue (upper panels). Mutations leading to classical Marfan syndrome are indicated as cMFS, and mutations leading to neonatal Marfan syndrome are indicated as nMFS. Note that attempts to obtain sequence information revealed nine degradation products as the N terminus of the respective proteins, and some sequences were not interpretable. Each lane of the degradation analyses was analyzed by densitometry and plotted as a three-dimensional graph (lower panels). The profile of the WT is depicted in black, the cMFS mutations are in light gray, and the nMFS mutations are in dark gray. High molecular weight bands are positioned on the left.
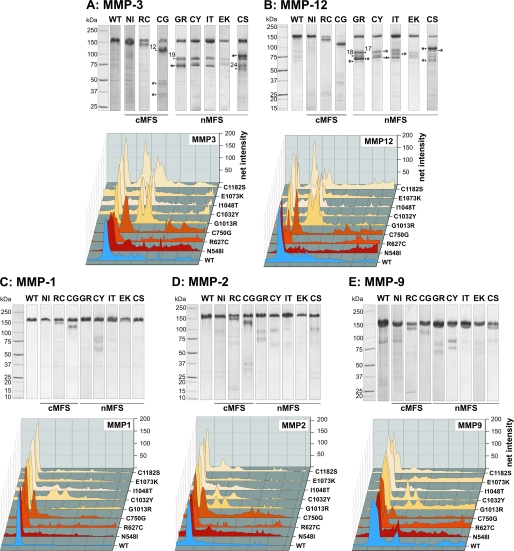
Matrix Metalloproteinases
The mutant proteins were digested with various MMPs as enhanced MMP expression has been shown in aortic specimens derived from patients with MFS (36). MMPs may play a role in the pathogenesis of MFS by degrading fibrillin and microfibrils, ultimately resulting in aortic dilatation, dissection, and rupture. In addition, it has been demonstrated that fibrillins are a direct target for certain MMPs (31, 32). The rF20 mutant proteins were susceptible for proteolytic attack by MMP-3 and -12 and to a much lesser extent by MMP-1, MMP-2, and MMP-9 (Fig. 6). Similar to the other proteases described above, MMP-3 and -12 primarily degraded all mutant proteins harboring nMFS mutations and the C750G cMFS mutant protein. Other cMFS mutations were less susceptible (Fig. 6, A and B). Typically, more degradation bands were observed for the nMFS mutant proteins versus the cMFS. Identified neo-cleavage sites reside in the mutated domain (G1013R, C1182S) or in the adjacent C-terminal domain (C750G) (Table 3).
FIGURE 6.
Proteolysis of rF20 polypeptides by MMPs. 7 μg of each polypeptide was digested with MMP-3 (A), MMP-12 (B), MMP-1 (C), MMP-2 (D), and MMP-9 (E). Degradation products were separated by SDS-PAGE under reducing conditions and stained with Coomassie Brilliant Blue (upper panels). The numbered arrows correlate with the determined N-terminal sequences of the degradation products summarized in Table 3. The abbreviations of mutations are used according to Table 1. Mutations leading to classical Marfan syndrome are indicated as cMFS, and mutations leading to neonatal Marfan syndrome are indicated as nMFS. Fragments with an N-terminal amino acid sequence of APLADYCQ (N terminus of rF20) are labeled with an asterisk (*). Each lane of the degradation analyses was analyzed by densitometry and plotted as a three-dimensional graph (lower panels). The profile of the WT is depicted in blue, the cMFS mutations is in shades of red, and the nMFS mutations are in shades of beige. High molecular weight bands are positioned on the left.
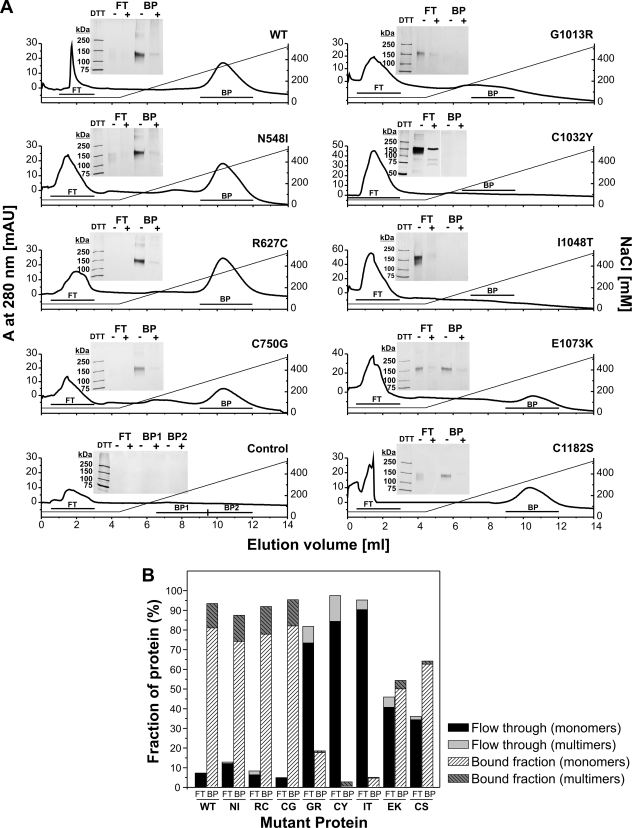
Functional Analysis of Mutant Fibrillin-1 Proteins: Heparin Interaction
One of the four heparin/heparan sulfate binding sites of fibrillin-1 is located in the neonatal region (Fig. 1A). Heparan sulfate is important for (i) microfibril assembly, (ii) perlecan-fibrillin interaction, and (iii) potentially for cell-matrix interactions (49, 55, 56). Therefore, we analyzed the heparin binding properties of the rF20 mutant proteins using heparin affinity chromatography (Fig. 7). Bound protein was eluted with a linear sodium chloride gradient, and pooled fractions were separated under reducing and non-reducing conditions by SDS-PAGE followed by silver staining and densitometry. All of the proteins containing cMFS mutations and the non-mutated rF20 bound to the column (Fig. 7A, left panel and Fig. 7B). There was no difference in the concentration of sodium chloride required for elution (200–400 mm), which is consistent with previous observations with smaller (non-mutated) fibrillin-1 fragments comprising the neonatal region only (49). These results indicate that the cMFS mutations did not affect the affinity of the proteins for heparin. The analysis of the proteins harboring nMFS mutations revealed that all mutations affected the proteins such that they did either not bind at all (G1013R, C1032Y, I1048T) or bound poorly (E1073K, C1182S) to the heparin column (Fig. 7, A, right panel, and B). In summary, the mutant proteins with nMFS mutations have lost or reduced their ability to interact with heparin.
FIGURE 7.
Interaction of mutant fibrillin-1 polypeptides with heparin. The mutant polypeptides were subjected to heparin affinity chromatography and eluted by a linear NaCl gradient as described under “Experimental Procedures.” A, binding and elution profiles at 280 nm are shown. Non-bound protein is detected in the flow-through (FT), and the bound protein (BP) is eluted by a NaCl gradient. The insets confirm the presence of the protein in the peaks after SDS-PAGE under reducing (DTT+) and non-reducing (DTT−) conditions and silver staining. Note that the silver-staining procedure stains reduced proteins to a lesser degree than non-reduced proteins. B, shown is quantification of the amount of bound and non-bound protein of the affinity chromatographies shown in A. The bars show the amount of monomers and multimers as indicated.
DISCUSSION
The mechanisms by which a mutation in the neonatal region of fibrillin-1 (TB3-cbEGF18) leads to severe nMFS remain unidentified. This study was designed to address potential pathogenetic mechanisms contributing to nMFS. We found that (i) neither selected cMFS nor nMFS mutations affected the overall stability of the protein, (ii) nMFS mutations generally resulted in a higher susceptibility for proteolytic digestion with some proteases, and (iii) nMFS mutations resulted in a loss of binding to heparin. Enhanced proteolytic susceptibility and interference with heparin binding could determine the severity of nMFS.
CD spectroscopy, which provides information about the average secondary structure of the mutant proteins, revealed no major changes in the relative amount of α-helices, β-sheets, and β-turns. In contrast, Mellody et al. (44) found significant structural changes by CD analysis of three cMFS mutations (G880S, C862R, and C908R) located in the second hybrid domain of fibrillin-1, which is contained in the rF20 fragment used in the present study. The recombinant fibrillin-1 fragment used in that study is considerable smaller (cbEGF7-cbEGF11) than the fragment used in the present study. Gross structural changes in a small fragment may appear as very small and local structural changes in a large fragment. It is also possible that MFS mutations in the second hybrid domain have more profound effects on the shape and the secondary structure of the molecule compared with mutations in other domains. None of the mutations analyzed in the present study is located in this domain. Heat stability analyzed by CD spectroscopy was also not markedly changed in the present study when comparing the control protein to the proteins containing the cMFS and nMFS mutations. The results emphasize that the mutations do not cause gross structural changes in the mutant proteins and demonstrate that this region of fibrillin-1 is structurally very stable. These data correlate with data obtained with smaller fibrillin-1 fragments harboring MFS mutations (13, 30). On the other hand, SDS-PAGE and gel filtration chromatography indicated a slightly faster mobility for C750G, I1048T, C1032Y, and C1182S compared with other mutants and to the control. Taken together, the structural analyses are compatible with the possibility that these mutant proteins were differently glycosylated despite a similar overall secondary structure. This hypothesis will be tested in an independent study. Overall, only small amounts of multimerization were observed for some of the mutant proteins, correlating with previous observations that fibrillin-1 multimerization is mediated through a C-terminal-located region (57).
To test for subtle structural changes in the fibrillin-1 mutant proteins, the non-physiological proteases trypsin and chymotrypsin were used to probe for cryptic cleavage sites exposed by the respective mutation. The use of these proteases is an established model for probing fibrillin stability, as in the calcium-saturated state recombinant fibrillin fragments are relatively resistant to proteolysis (13, 14, 58). The large number of predicted cleavage sites allows for the fine mapping of mutational effects. Physiological proteases (MMPs, thrombin, plasmin, cathepsins) were included in this study because they are actively involved in matrix remodeling and pathological matrix degradation. Several of them have been described as being up-regulated in aortic tissue from patients with Marfan syndrome (33, 36). In contrast to trypsin and chymotrypsin, only a few cleavage sites are predicted in fibrillin-1 for these physiological proteases.
The rF20 polypeptides with nMFS mutations were clearly more susceptible to trypsin and chymotrypsin, indicating that nMFS mutations resulted in more extensive structural disturbances compared with cMFS mutations. Neonatal constructs were also more susceptible to cathepsins K and V, MMP-3, MMP-12, and to a lesser degree, plasmin. MMP-1,-2, and -9 had little overall effect on the degradation of rF20-WT and the mutant polypeptides despite that MMP-2 and -9 have been described as proteases involved in the pathogenesis of aortic aneurysms (33, 36). MMP-9 and -12 cleavage sites implicated in (non-mutated) fibrillin-1 degradation are not located within rF20 but were localized to the junction between the proline-rich region and EGF4 (MMP-9) and at the C-terminal end of cbEGF31 (for MMP-12) (31). In our study, however, we found major MMP-12 neo-cleavage sites in the TB3 domain of the rF20 fragment harboring the nMFS mutation G1013R. This indicates that not all the potential cleavage sites for MMP-12 in fibrillin-1 are accessible under normal conditions and that mutations may expose major cleavage sites for MMP-12. More generally, it appears that most or all of the cleavage sites observed in this study after the digestion of the mutant proteins with MMP-3 and -12 occurred as a consequence of the mutation exposing cryptic cleavage sites, as only very few cleavage products were observed for the rF20-WT fragment.
Cathepsin K and V are highly active elastases (37, 39, 59). Here, we demonstrate for the first time that these cathepsins are able to degrade fibrillin-1 using recombinant non-mutated fibrillin-1 fragments spanning the entire molecule. Mapping of the cleavage sites indicates that these cathepsins exclusively cleave fibrillin-1 in linker regions between EGF1 and EGF2, the proline-rich region and EGF4, TB2 and cbEGF7, and TB6 and cbEGF32. These inter-domain cleavage sites likely have more severe consequences on the stability of microfibrils than intra-domain cleavage sites, where disulfide bonds still hold the cleaved fragments together. Generally, mutant fibrillin-1 proteins harboring nMFS mutations showed enhanced proteolytic degradation patterns after treatment with cathepsins K and V. These data suggest that both cathepsins may play a role in the pathogenesis of nMFS.
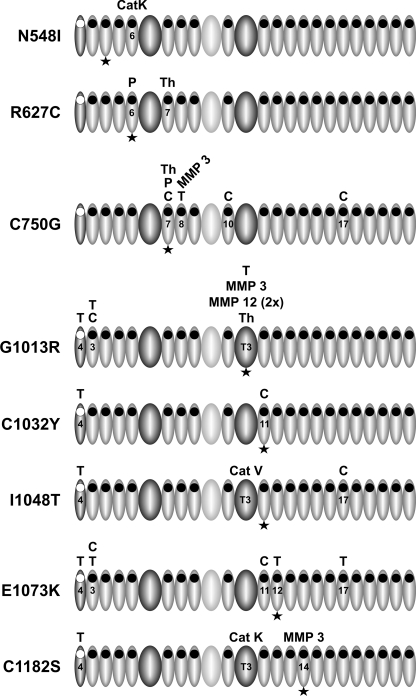
We mapped protease cleavage sites by N-terminal sequencing to analyze how the mutations affected protease susceptibility of the polypeptides and to determine neo-cleavage sites for the proteases. Most of the neo-cleavage sites were observed close to the mutation site either directly within the mutated domain or in the adjacent domains, which is in line with previous publications by us and others (Fig. 8) (12, 13, 16, 30, 58). However, some of the cleavage sites were found several domains away from the mutation site. These long range effects could not be predicted with smaller fragments used in previous studies (13, 15, 58, 60). We speculate that those long range effects are caused consecutively after the initial cleavage sites had occurred relatively close to the mutations. These initial cleavage sites may trigger long range structural modifications. In tissues, fibrillins may be exposed to a complex mixture of proteases so that cleavage and degradation of mutant fibrillin and microfibrils may involve subsequent events. Cleavage by enzyme A may result in the exposure of cleavage sites for enzyme B. Initial efforts, however, to resolve the sequence of cleavage events were not successful because the time resolution of our experimental setup is currently insufficient (data not shown).
FIGURE 8.
Schematic representation of the position of neo-cleavage sites in rF20 mutant proteins. The figure depicts graphically the data presented in Table 3 for each mutation separately. Only the domains for which neo-cleavage sites were identified are numbered. The domains are coded as described in Fig. 1. The position of the mutation is indicated with an asterisk. T, trypsin; C, chymotrypsin; P, plasmin; Th, thrombin; MMP, matrix metalloproteinase; Cat, cathepsin.
The region Pro1009–Lys1027, representing the linker between TB3 and cbEGF11 at the N-terminal side of the neonatal region, was found to be a very sensitive proteolytic site (Table 3). A recent study found a higher probability of nMFS associated with mutations in the adjacent cbEGF11 (Asp1028–Thr1069) as well as a higher probability of cardiovascular manifestations such as ascending aortic dilatation and mitral valve regurgitation as compared with other cbEGF domains within the neonatal region (17). Patients with a mutation in cbEGF11 were diagnosed with nMFS earlier than patients with mutations elsewhere in the neonatal region. The rate of survival was also markedly reduced in patients with mutations in cbEGF11. Consequently, this region is a significant site that can contribute to the pathogenesis of nMFS, and cleavage in the loop connecting TB3 to cbEGF11 may contribute to the pathogenetic mechanism.
To analyze functional consequences of the mutations, we determined interaction of the mutant fibrillin-1 polypeptides with heparin, one of the known and experimentally accessible binding partners contained within the rF20 fragment. Interactions of fibrillin-1 with heparin/heparan sulfate-containing proteoglycans have been suggested to play a critical role in microfibril assembly (49, 61). All the neonatal mutations either lost (G1013R, C1032Y, I1048T) or had a reduced capability (E1073K, C1182S) to bind to heparin, whereas none of the classical mutations had a negative effect on heparin interaction. It is remarkable that single nMFS point mutations are sufficient for a complete inability of the mutant proteins to interact with heparin. Some mutations, such as G1013R and E1073K, located in the heparin binding region may exert their negative effect on heparin binding through modification of surface charges. However, because enhanced proteolysis indicated local and, in some cases, long range structural changes, we propose that such structural changes are responsible for the loss of heparin binding. When comparing the effects of nMFS and cMFS mutations on heparin binding, a limitation of the study is the fact that all the cMFS mutations are located further upstream of the heparin binding site, whereas the nMFS mutations are located closer to the heparin binding site. Therefore, potential short and longer range structural effects induced by the cMFS mutations may not reach the heparin binding site or may not be large enough to interfere with heparin binding.
In summary, the present study suggests that enhanced proteolytic susceptibility, especially in the linker region between TB3 and cbEGF11, and functional loss of the central fibrillin-1 heparin/heparan sulfate interactions contribute to the development of the more severe nMFS as compared with cMFS.
Supplementary Material
This work was supported by the Canadian Institutes of Health Research Grants IMH-102821 and MOP-106494, the Canadian Marfan Association, and the Canada Foundation for Innovation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2.
- MFS
- Marfan syndrome
- cMFS
- classical Marfan syndrome
- nMFS
- neonatal Marfan syndrome
- MMP
- matrix metalloproteinase
- BCA
- bichinonic acid
- cbEGF
- calcium-binding EGF-like
- TB
- TGF-β-binding protein like
- NI
- N548I
- RC
- R627C
- CG
- C750G
- GR
- G1013R
- CY
- C1032Y
- IT
- I1048T
- EK
- E1073K
- CS
- C1182S.
REFERENCES
- 1. Robinson P. N., Arteaga-Solis E., Baldock C., Collod-Béroud G., Booms P., De Paepe A., Dietz H. C., Guo G., Handford P. A., Judge D. P., Kielty C. M., Loeys B., Milewicz D. M., Ney A., Ramirez F., Reinhardt D. P., Tiedemann K., Whiteman P., Godfrey M. (2006) J. Med. Genet. 43, 769–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Loeys B. L., Gerber E. E., Riegert-Johnson D., Iqbal S., Whiteman P., McConnell V., Chillakuri C. R., Macaya D., Coucke P. J., De Paepe A., Judge D. P., Wigley F., Davis E. C., Mardon H. J., Handford P., Keene D. R., Sakai L. Y., Dietz H. C. (2010) Sc. Transl. Med. 2, 23ra20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ramirez F., Rifkin D. B. (2009) Curr. Opin. Cell Biol. 21, 616–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hubmacher D., Tiedemann K., Reinhardt D. P. (2006) Curr. Top. Dev. Biol. 75, 93–123 [DOI] [PubMed] [Google Scholar]
- 5. Kielty C. M., Sherratt M. J., Marson A., Baldock C. (2005) Adv. Protein Chem. 70, 405–436 [DOI] [PubMed] [Google Scholar]
- 6. Handford P., Downing A. K., Rao Z., Hewett D. R., Sykes B. C., Kielty C. M. (1995) J. Biol. Chem. 270, 6751–6756 [DOI] [PubMed] [Google Scholar]
- 7. Yuan X., Downing A. K., Knott V., Handford P. A. (1997) EMBO J. 16, 6659–6666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smallridge R. S., Whiteman P., Werner J. M., Campbell I. D., Handford P. A., Downing A. K. (2003) J. Biol. Chem. 278, 12199–12206 [DOI] [PubMed] [Google Scholar]
- 9. Wagenseil J. E., Mecham R. P. (2007) Birth Defects Res. C. Embryo. Today 81, 229–240 [DOI] [PubMed] [Google Scholar]
- 10. Pyeritz R. E. (2000) Annu. Rev. Med. 51, 481–510 [DOI] [PubMed] [Google Scholar]
- 11. Collod-Béroud G., Le Bourdelles S., Ades L., Ala-Kokko L., Booms P., Boxer M., Child A., Comeglio P., De Paepe A., Hyland J. C., Holman K., Kaitila I., Loeys B., Matyas G., Nuytinck L., Peltonen L., Rantamaki T., Robinson P., Steinmann B., Junien C., Béroud C., Boileau C. (2003) Hum. Mutat. 22, 199–208 [DOI] [PubMed] [Google Scholar]
- 12. Suk J. Y., Jensen S., McGettrick A., Willis A. C., Whiteman P., Redfield C., Handford P. A. (2004) J. Biol. Chem. 279, 51258–51265 [DOI] [PubMed] [Google Scholar]
- 13. Vollbrandt T., Tiedemann K., El-Hallous E., Lin G., Brinckmann J., John H., Bätge B., Notbohm H., Reinhardt D. P. (2004) J. Biol. Chem. 279, 32924–32931 [DOI] [PubMed] [Google Scholar]
- 14. Reinhardt D. P., Ono R. N., Sakai L. Y. (1997) J. Biol. Chem. 272, 1231–1236 [DOI] [PubMed] [Google Scholar]
- 15. Kettle S., Yuan X., Grundy G., Knott V., Downing A. K., Handford P. A. (1999) J. Mol. Biol. 285, 1277–1287 [DOI] [PubMed] [Google Scholar]
- 16. McGettrick A. J., Knott V., Willis A., Handford P. A. (2000) Hum. Mol. Genet. 9, 1987–1994 [DOI] [PubMed] [Google Scholar]
- 17. Faivre L., Collod-Beroud G., Callewaert B., Child A., Binquet C., Gautier E., Loeys B. L., Arbustini E., Mayer K., Arslan-Kirchner M., Stheneur C., Kiotsekoglou A., Comeglio P., Marziliano N., Wolf J. E., Bouchot O., Khau-Van-Kien P., Beroud C., Claustres M., Bonithon-Kopp C., Robinson P. N., Adès L., De, Backer J., Coucke P., Francke U., De Paepe A., Jondeau G., Boileau C. (2009) Eur. J. Hum. Genet. 17, 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Neptune E. R., Frischmeyer P. A., Arking D. E., Myers L., Bunton T. E., Gayraud B., Ramirez F., Sakai L. Y., Dietz H. C. (2003) Nat. Genet. 33, 407–411 [DOI] [PubMed] [Google Scholar]
- 19. Brooke B. S., Habashi J. P., Judge D. P., Patel N., Loeys B., Dietz H. C., 3rd (2008) N. Engl. J. Med. 358, 2787–2795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ng C. M., Cheng A., Myers L. A., Martinez-Murillo F., Jie C., Bedja D., Gabrielson K. L., Hausladen J. M., Mecham R. P., Judge D. P., Dietz H. C. (2004) J. Clin. Invest. 114, 1586–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Milewicz D. M., Pyeritz R. E., Crawford E. S., Byers P. H. (1992) J. Clin. Invest. 89, 79–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aoyama T., Tynan K., Dietz H. C., Francke U., Furthmayr H. (1993) Hum. Mol. Genet. 2, 2135–2140 [DOI] [PubMed] [Google Scholar]
- 23. Hollister D. W., Godfrey M., Sakai L. Y., Pyeritz R. E. (1990) N. Engl. J. Med. 323, 152–159 [DOI] [PubMed] [Google Scholar]
- 24. Raghunath M., Superti-Furga A., Godfrey M., Steinmann B. (1993) Hum. Genet. 90, 511–515 [DOI] [PubMed] [Google Scholar]
- 25. Kielty C. M., Phillips J. E., Child A. H., Pope F. M., Shuttleworth C. A. (1994) Matrix Biol. 14, 191–199 [DOI] [PubMed] [Google Scholar]
- 26. Raghunath M., Kielty C. M., Kainulainen K., Child A., Peltonen L., Steinmann B. (1994) Biochem. J. 302, 889–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Halme T., Savunen T., Aho H., Vihersaari T., Penttinen R. (1985) Exp. Mol. Pathol. 43, 1–12 [DOI] [PubMed] [Google Scholar]
- 28. Fleischer K. J., Nousari H. C., Anhalt G. J., Stone C. D., Laschinger J. C. (1997) Ann. Thorac. Surg. 63, 1012–1017 [DOI] [PubMed] [Google Scholar]
- 29. Tsuji T. (1986) J. Cutan. Pathol. 13, 144–153 [DOI] [PubMed] [Google Scholar]
- 30. Reinhardt D. P., Ono R. N., Notbohm H., Müller P. K., Bächinger H. P., Sakai L. Y. (2000) J. Biol. Chem. 275, 12339–12345 [DOI] [PubMed] [Google Scholar]
- 31. Hindson V. J., Ashworth J. L., Rock M. J., Cunliffe S., Shuttleworth C. A., Kielty C. M. (1999) FEBS Lett. 452, 195–198 [DOI] [PubMed] [Google Scholar]
- 32. Ashworth J. L., Murphy G., Rock M. J., Sherratt M. J., Shapiro S. D., Shuttleworth C. A., Kielty C. M. (1999) Biochem. J. 340, 171–181 [PMC free article] [PubMed] [Google Scholar]
- 33. Ikonomidis J. S., Jones J. A., Barbour J. R., Stroud R. E., Clark L. L., Kaplan B. S., Zeeshan A., Bavaria J. E., Gorman J. H., 3rd, Spinale F. G., Gorman R. C. (2006) Circulation 114, I365-I370 [DOI] [PubMed] [Google Scholar]
- 34. Chung A. W., Au Yeung K., Sandor G. G., Judge D. P., Dietz H. C., van Breemen C. (2007) Circ. Res. 101, 512–522 [DOI] [PubMed] [Google Scholar]
- 35. Booms P., Pregla R., Ney A., Barthel F., Reinhardt D. P., Pletschacher A., Mundlos S., Robinson P. N. (2005) Hum. Genet. 116, 51–61 [DOI] [PubMed] [Google Scholar]
- 36. Segura A. M., Luna R. E., Horiba K., Stetler-Stevenson W. G., McAllister H. A., Jr., Willerson J. T., Ferrans V. J. (1998) Circulation 98, II331-II337 [PubMed] [Google Scholar]
- 37. Yasuda Y., Li Z., Greenbaum D., Bogyo M., Weber E., Brömme D. (2004) J. Biol. Chem. 279, 36761–36770 [DOI] [PubMed] [Google Scholar]
- 38. Wilson S. R., Peters C., Saftig P., Brömme D. (2009) J. Biol. Chem. 284, 2584–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Samokhin A. O., Wong A., Saftig P., Brömme D. (2008) Atherosclerosis 200, 58–68 [DOI] [PubMed] [Google Scholar]
- 40. Chaudhry S. S., Cain S. A., Morgan A., Dallas S. L., Shuttleworth C. A., Kielty C. M. (2007) J. Cell Biol. 176, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lin G., Tiedemann K., Vollbrandt T., Peters H., Batge B., Brinckmann J., Reinhardt D. P. (2002) J. Biol. Chem. 277, 50795–50804 [DOI] [PubMed] [Google Scholar]
- 42. Jensen S. A., Reinhardt D. P., Gibson M. A., Weiss A. S. (2001) J. Biol. Chem. 276, 39661–39666 [DOI] [PubMed] [Google Scholar]
- 43. Reinhardt D. P., Keene D. R., Corson G. M., Pöschl E., Bächinger H. P., Gambee J. E., Sakai L. Y. (1996) J. Mol. Biol. 258, 104–116 [DOI] [PubMed] [Google Scholar]
- 44. Mellody K. T., Freeman L. J., Baldock C., Jowitt T. A., Siegler V., Raynal B. D., Cain S. A., Wess T. J., Shuttleworth C. A., Kielty C. M. (2006) J. Biol. Chem. 281, 31854–31862 [DOI] [PubMed] [Google Scholar]
- 45. Provencher S. W., Glöckner J. (1981) Biochemistry 20, 33–37 [DOI] [PubMed] [Google Scholar]
- 46. van Stokkum I. H., Spoelder H. J., Bloemendal M., van Grondelle R., Groen F. C. (1990) Anal. Biochem. 191, 110–118 [DOI] [PubMed] [Google Scholar]
- 47. Linnevers C. J., McGrath M. E., Armstrong R., Mistry F. R., Barnes M. G., Klaus J. L., Palmer J. T., Katz B. A., Brömme D. (1997) Protein Sci. 6, 919–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brömme D., Li Z., Barnes M., Mehler E. (1999) Biochemistry 38, 2377–2385 [DOI] [PubMed] [Google Scholar]
- 49. Tiedemann K., Bätge B., Müller P. K., Reinhardt D. P. (2001) J. Biol. Chem. 276, 36035–36042 [DOI] [PubMed] [Google Scholar]
- 50. Abramoff M. D., Magelhaes P. J., Ram S. J. (2004) Biophoton. Int. 11, 36–42 [Google Scholar]
- 51. Shang J. (2005) Methods 35, 390–394 [DOI] [PubMed] [Google Scholar]
- 52. Smallridge R. S., Whiteman P., Doering K., Handford P. A., Downing A. K. (1999) J. Mol. Biol. 286, 661–668 [DOI] [PubMed] [Google Scholar]
- 53. Werner J. M., Knott V., Handford P. A., Campbell I. D., Downing A. K. (2000) J. Mol. Biol. 296, 1065–1078 [DOI] [PubMed] [Google Scholar]
- 54. Lutgens E., Lutgens S. P., Faber B. C., Heeneman S., Gijbels M. M., de Winther M. P., Frederik P., van der Made I, Daugherty A., Sijbers A. M., Fisher A., Long C. J., Saftig P., Black D., Daemen M. J., Cleutjens K. B. (2006) Circulation 113, 98–107 [DOI] [PubMed] [Google Scholar]
- 55. Cain S. A., Baldock C., Gallagher J., Morgan A., Bax D. V., Weiss A. S., Shuttleworth C. A., Kielty C. M. (2005) J. Biol. Chem. 280, 30526–30537 [DOI] [PubMed] [Google Scholar]
- 56. Tiedemann K., Sasaki T., Gustafsson E., Göhring W., Bätge B., Notbohm H., Timpl R., Wedel T., Schlötzer-Schrehardt U., Reinhardt D. P. (2005) J. Biol. Chem. 280, 11404–11412 [DOI] [PubMed] [Google Scholar]
- 57. Hubmacher D., El-Hallous E. I., Nelea V., Kaartinen M. T., Lee E. R., Reinhardt D. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6548–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Whiteman P., Smallridge R. S., Knott V., Cordle J. J., Downing A. K., Handford P. A. (2001) J. Biol. Chem. 276, 17156–17162 [DOI] [PubMed] [Google Scholar]
- 59. Bühling F., Waldburg N., Gerber A., Häckel C., Krüger S., Reinhold D., Brömme D., Weber E., Ansorge S., Welte T. (2000) Adv. Exp. Med. Biol. 477, 281–286 [DOI] [PubMed] [Google Scholar]
- 60. Yuan X., Werner J. M., Lack J., Knott V., Handford P. A., Campbell I. D., Downing A. K. (2002) J. Mol. Biol. 316, 113–125 [DOI] [PubMed] [Google Scholar]
- 61. Ritty T. M., Broekelmann T. J., Werneck C. C., Mecham R. P. (2003) Biochem. J. 375, 425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dietz H. C., McIntosh I., Sakai L. Y., Corson G. M., Chalberg S. C., Pyeritz R. E., Francomano C. A. (1993) Genomics 17, 468–475 [DOI] [PubMed] [Google Scholar]
- 63. Hayward C., Rae A. L., Porteous M. E., Logie L. J., Brock D. J. (1994) Hum. Mol. Genet. 3, 373–375 [DOI] [PubMed] [Google Scholar]
- 64. Nijbroek G., Sood S., McIntosh I., Francomano C. A., Bull E., Pereira L., Ramirez F., Pyeritz R. E., Dietz H. C. (1995) Am. J. Hum. Genet. 57, 8–21 [PMC free article] [PubMed] [Google Scholar]
- 65. Ng D. K., Chau K. W., Black C., Thomas T. M., Mak K. L., Boxer M. (1999) J. Paediatr. Child Health 35, 321–323 [DOI] [PubMed] [Google Scholar]
- 66. Perez A. B., Pereira L. V., Brunoni D., Zatz M., Passos-Bueno M. R. (1999) Hum. Mutat. 13, 84 [DOI] [PubMed] [Google Scholar]
- 67. Brune D. C. (1992) Anal. Biochem. 207, 285–290 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.