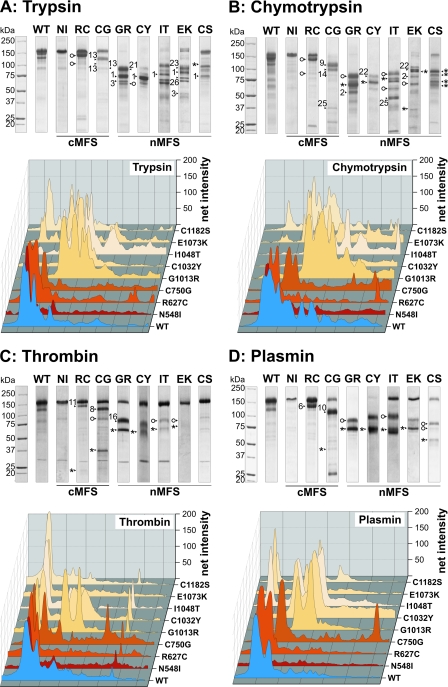
FIGURE 4.
Proteolysis of rF20 polypeptides with various non-physiological and physiological proteases. 7 μg of each polypeptide was digested with trypsin (A), chymotrypsin (B), thrombin (C), and plasmin (D) as detailed under “Experimental Procedures.” Proteolytic degradation products were separated by SDS-PAGE under reducing conditions and stained with Coomassie Brilliant Blue (upper panels). The numbered arrows correlate with the determined N-terminal sequences of the degradation products summarized in Table 3. The abbreviations of mutations are used according to Table 1. Mutations leading to classical Marfan syndrome are indicated as cMFS, and mutations leading to neonatal Marfan syndrome are indicated as nMFS. Fragments starting with the “APLADYCQ” amino acid sequence (N terminus of rF20) are labeled with an asterisk (*). Cleavage sites marked with an open circle were also found in the wild-type rF20 (WT). The lanes for each of the degradation analyses were analyzed by densitometry and plotted as a three-dimensional graph (lower panels). The profile of the WT is depicted in blue, the cMFS mutations is in shades of red, and the nMFS mutations are in shades of beige. High molecular weight bands are positioned on the left.