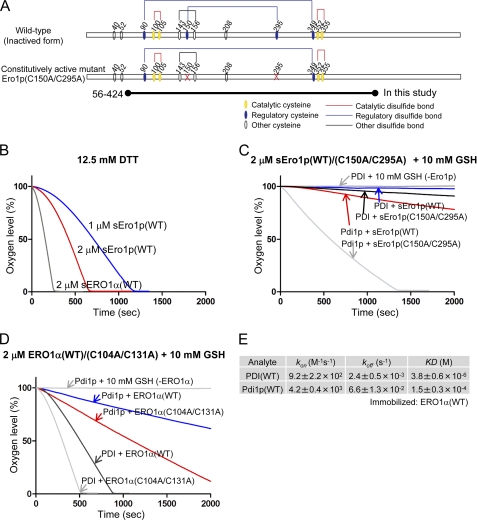
FIGURE 3.
In vitro complementation assay between the human PDI-ERO1α and yeast Pdi1p-Ero1p pathway. A, wild-type and constitutively active mutant of yeast Ero1p. Cysteine residues are shown with circles. Structural, regulatory, and catalytic disulfides are represented by black, blue, and red lines, respectively (7). The short form of yeast Ero1p, spanning amino acid residues 56–424, was examined; it is named sEro1p (12, 21). B, oxygen consumption by yeast sEro1p or human ERO1α oxidation of 12.5 mm DTT. Kinetics of oxygen consumption by 2 μm yeast wild-type or constitutively active sEro1p (C), or 2 μm human wild-type or constitutively active ERO1α (D), during the reaction with 5 μm yeast Pdi1p or human PDI in the presence of GSH (10 mm). E, calculated kinetic parameters for affinity measurements between human PDI and ERO1α(WT) or yeast Pdi1p and ERO1α(WT) by SPR spectroscopy. Sensorgrams are shown in supplemental Fig. S2.