Figure 4.
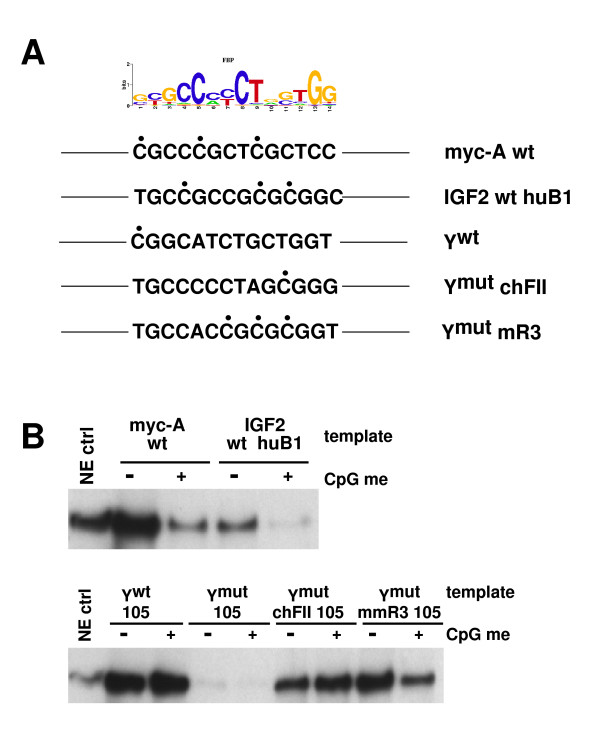
Cytosine methylation of the CTCF core motif Y does not influence binding of CTCF. (A) CTCF motifs used in the context of the 105-bp immobilized template derived from the intronic region of the IGF2BP1 gene are shown. The position frequency matrix of the CTCF target motif is shown at the top. Only the sense strand of the motifs is shown. CpG residues are indicated by filled black circles. Myc-A, IGF2 huB1 and Ywt are CTCF target sequences derived from MYC, IGF2 and IGF2BP1 gene loci. Ymut chFII and Ymut mmR3 contain the CTCF target sequence of the chicken HS4 insulator [57] and the CTCF target region of the mouse imprinting control region R3 [10]. (B) Top: control experiments revealed the sensitivity of CTCF binding to DNA methylation (CpG me) at the myc-A and IGF2 huB1 templates. Bottom: methylation of the 105-bp Ywt template did not affect the recruitment of CTCF. While methylated chicken FII CTCF target sites efficiently recruited CTCF, CpG methylation of the mouse R3 sequence decreased the binding of CTCF.