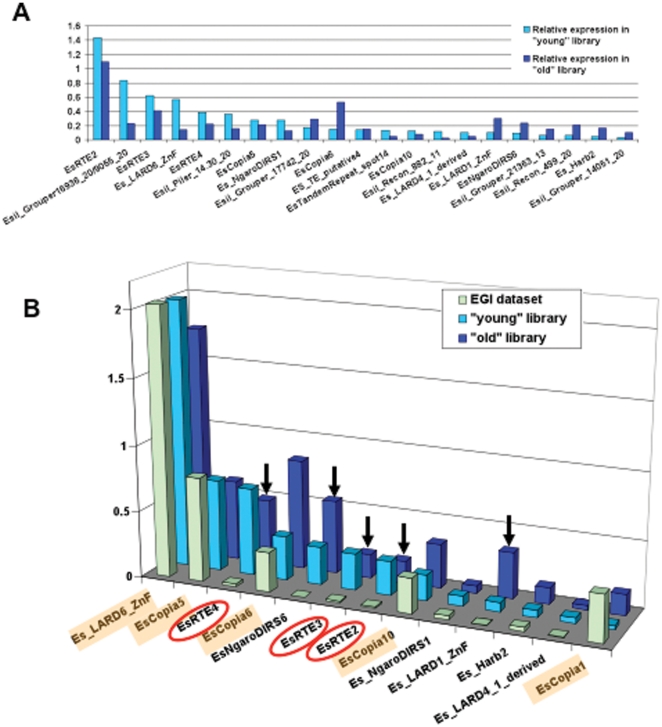
Figure 3. Over-representation of Ectocarpus transposable elements (TEs) in Eurychasma-infected libraries.
A. Relative expression of Ectocarpus TEs in the pyrosequenced “young” and “old” Eurychasma-infected libraries. Total read counts were normalised over contig length in order to identify the most expressed unigenes in each library. Expression values are given for all TEs belonging to the top 150 most highly expressed unigenes in at least one library. If several contigs matched the same TE in a given library, their relative expression levels were summed. TE nomenclature is as per Maumus (2009). B. Differential expression pattern of Ectocarpus TEs between the pyrosequenced “young” and “old” Eurychasma-infected libraries vs. the Ectocarpus Genome Initiative EST collection (“EGI dataset”, Cock et al, 2010). For each library, sequence read counts were normalised over the genome coverage of each TE in order to control for the leaky transcription hypothesis. The 5 most highly expressed TEs in the EGI dataset are highlighted in orange; those circled in red belong to the 10 most abundant repeated elements identified in the Ectocarpus genome. Arrows point to TEs highly induced in the “young” and “old” infected cultures.