Abstract
MMP19 and MMP23B belong to the Matrix metalloproteases (MMPs) family, which are zinc-binding endopeptidases that are capable of degrading various components of the extracellular matrix. They are thought to play important roles in embryonic development, reproduction and tissue remodeling, as well as in cell proliferation, differentiation, migration, angiogenesis, apoptosis and host defense. However, they are poorly understood in pigs. Here, we obtained the full length coding region sequence and genomic sequence of the porcine MMP19 and MMP23B genes and analyzed their genomic structures. The deduced amino acid sequence shares similar precursor protein domains with human and mouse MMP19 and MMP23B protein, respectively. Using IMpRH panel, MMP19 was mapped to SSC5p12-q11 (closely linked to microsatellite DK) and MMP23B was mapped to SSC8q11-q12 (linked to microsatellite Sw2521). Quantitative real-time PCR showed that MMP19 was abundantly expressed in the liver, while MMP23B was strongly expressed in the ovarian and heart. Furthermore, both genes were all expressed increasingly in prenatal skeletal muscle during development. Three SNPs were detected by sequencing and PCR-RFLP methods, and association analysis indicated that C203T at exon 5 of MMP19 has a significant association with the blood parameters WBC (G/L) and IgG2 (mg/mL) (P<0.05), SNP C131T at exon 3 of MMP23B is significantly associated with the blood parameters HGB (g/L) and MCH (P<0.05), and A150G in exon 4 has no significant association with the economic traits in pigs.
Keywords: pig, MMP19, MMP23B, chromosome mapping, expression, association analysis.
Introduction
Matrix metalloproteinases (MMPs), also called matrixins, are zinc-dependent endopeptidases that are involved in extracellular matrix (ECM) degradation by degrading a number of extracellular molecules and bioactive molecules, thereby playing a central role in normal physiological processes, such as embryonic development, reproduction, and tissue remodeling, as well as in cell proliferation, migration, differentiation, angiogenesis, apoptosis and host defenses. MMP19 and MMP23B are members of the Matrix metalloproteinases (MMPs) family.
The MMP19 gene was cloned from a rheumatoid arthritis patient 1 and human liver cDNA library 2. It is expressed widely in normal human tissues, including the mammary gland, placenta, lung, pancreas, ovarian, and other tissues 2. Additionally, MMP19 promotes proliferation, migration, and cell adhesion of keratinocytes 3-4, and it is a likely candidate to be the major IGFBP-3 degrading MMP in the quiescent epidermis because IGFBP-3 and MMP19 are both expressed in the skin 5. Since it is expressed in theca cells and granulosa cells, MMP19 is suggested that functions during follicular growth, ovulation, and luteal regression 5-6. Moreover, MMP19 is highly expressed in proliferating astrocytoma/glioma cells, and its expression may facilitate the cell's invasion through brain extracellular matrix components 7. In MDA-MB-231 cells, the decreased invasiveness appeared to be mediated by decreased transcript levels of MMP19 8. MMP19 also contributes to the integrin switch favoring epithelial migration and actively participates in the early stages of squamous cell cancer invasion 9. Knock-out mice deficient in MMP19 expression are viable and fertile and do not display any obvious abnormalities. However, they exhibit decreased susceptibility to skin tumors and adipocyte hypertrophy induced by diet 10. Based on the phenotype of MMP19-deficient mice, researchers proposed that MMP19 is an important factor in cutaneous immune responses and influences the development of T cells 11.
The human cDNA encoding MMP23 was cloned from an ovarian cDNA library and mapped to chromosome 1p36. It is expressed predominantly in ovarian, testis and prostate tissues 12. The rat homolog was cloned from gonadotropin-primed immature rat ovaries, and its expression is spatially and temporally regulated in a cell type-specific manner during follicular development 13. MMP23 was transcribed in chondrocytes and osteoblasts, suggesting a role in some aspect of cartilage or bone formation 14. In addition, MMP23 is expressed in the synovium and cartilage 15, at the cranial sutures 16 and in human amniochorion 17. And MMP23 is up-regulated in common bile duct ligation (CBDL) livers and therefore may be involved in hepatic bone marrow beta 2m(-)/Thy-1+ hepatic stem cells (BMHSCs) priming 18.
Current studies indicate that MMP19 promotes proliferation, migration and cell adhesion and influences immunity, while MMP23 may play important roles in follicular development and bone formation. However, little is known about the porcine MMP19 and MMP23 genes. To characterize the structures of porcine MMP19 and MMP23 genes and explore their possible functions in pig, multiple approaches were undertaken in our study. Porcine MMP19 and MMP23B genes were firstly cloned and sequenced. Sequently, the genes were mapped using IMpRH panel. And then, quantitative real-time PCR was employed to analyze the temporal and spatial expression in different tissues and skeletal muscle at different periods. Finally, SNP detection and association analyses were undertaken to discover the relationship between the genetic variation and the economic traits of pigs.
Materials and Methods
Animal samples and traits data collection
Twenty-two tissues, including heart, liver, spleen, kidney, small intestine, ovarian, abdomen fat, oviduct, large intestine, limb muscle and longissimus dorsi muscle from adult Landrace pigs and heart, liver, spleen, kidney, small intestine, ovarian, abdomen fat, back fat, lung, tongue and stomach from adult indigenous Chinese Tongcheng pigs, were collected for spatial expression analysis. Prenatal skeletal muscle was collected at three developmental stages (33, 65 and 90 days post conception (dpc)) from pregnant Landrace and Tongcheng pigs. All the tissue samples were harvested, immediately frozen in liquid nitrogen, and then stored at -80ºC until used. Unrelated individuals from 4 pure pig breeds (Landrace (n=3), Yorkshire (n=3), Tongcheng (n=3) and Wuzhishan (n=3)) were used to scan single-nucleotide polymorphism (SNP) sites. To identify variants, allele frequencies were estimated in a population composed of unrelated, randomly selected individuals representing Landrace, Yorkshire, Tongcheng and Laiwu pigs, as well as Wuzhishan, Bama and Guizhou mini pigs. The population used for association analysis contained 3 pure pig breeds (Landrace, Yorkshire and Tongcheng) and a further pair of 3-cross breeds: Landrace× (Large White × Tongcheng) (LYT) and Yorkshire × (Landrace × Tongcheng) (YLT) 19. Blood samples and traits data from individuals were obtained using standard methods, as described by Wang et al 20.
A total of 22 traits were measured. These traits included growth (average daily gain from birth to marketing, ADG), carcass (dressing percentage, DP; loin-muscle height, LH; backfat at the loin, XBF; eye muscle area, EMA; average backfat thickness at 3 points, BF; percentage of leaf and caul fat, PLCF; percentage of ham in the carcass, PHC), meat quality (meat color score, MCS; mobling score, MS; muscle tender, MT; pH value at 45-60 min postmortem, pH45; muscle shear force, MSF; intramuscular fat content, IMF) and immune traits (white blood cell counts, WBC; hemoglobin, HGB; total erythrocytes, RBC; mean corpuscular volume hemoglobin, MCH; mean corpuscular volume hemoglobin concentration, MCHC; hematocrit, HCT; red cell distribution width, RDW; concentration of IgG, IgG2).
Cloning of porcine MMP19 and MMP23B genes
The porcine MMP19 and MMP23B genes were isolated using an in silicon cloning strategy. The human MMP19 and MMP23B transcripts (NM_002429 and NM_006983, respectively) retrieved from the GenBank database (http://www.ncbi.nlm.nih.gov/) were used as entries to search for homologous sequence in the pig EST database (http://www.ncbi.nlm.nih.gov/genbank/GenbankSearch.html). The homologous EST sequence fragments showing above 85% identity and having a size of more than 100 bp were obtained and assembled using the SeqMan program (DNAStar, Inc., Madison, WI, USA). Primer pairs were designed from the assembled EST contigs with Primer 5.0 software to amplify porcine MMP19 and MMP23B cDNA sequences.
The PCR 50 µL reaction consisted of 100 ng DNA or cDNA template, 5 µL 10x PCR buffer, 4 µL 2.5 mM dNTP mix, 1 µL sense primer (100 ng/µL), 1 µL antisense primer (100 ng/µL), 0.5 µL 5 U r-Taq DNA polymerase (Fermentas, Vilnius, Lithuania) and sterile water. The PCR amplification was initiated with a 5-minute denaturation at 95°C and continued with the following parameters (35 cycles): 30 sec at 95°C (denaturation), 30 sec at Tm specific for primer (annealing) and 30 sec at 72°C (elongation), followed by 5 min at 72°C (elongation), and stored at 4°C until analysis. Rapid Amplification of cDNA Ends (RACE) was then employed to sequence the whole gene, as described in the user manual for RACE kit (Takara, Dalian, China). The total RNA and genomic DNA used for gene cloning were extracted from the blood samples of Wuzhishan mini pigs and the primer pairs used for PCR amplification are listed in Table 1.
Table 1.
Primers used in this study
| Gene | Primer symbol | Primer sequence (5'-3') | Tm (℃) |
Used for |
|---|---|---|---|---|
| MMP19 | MMP19.L | ACAGGGTCCGTATGAGGCA | 65 | cDNA cloning |
| MMP19 | MMP19.R | GGAAATCCGAGGCTCAACG | ||
| MMP19 | 5'RACE.MMP19.R | GGTTCAAGATGCGGAAGGTCAGGTGC | ||
| MMP19 | 3'RACE.MMP19.L | CCACGCCGTTGGATGGGGATACTTC | ||
| MMP19 | 3'RACE nest | TCTCCACCACCCACTCATTC | ||
| MMP23B | MMP23B-CDS.L | GACGCCGCTACACGCTGACC | 65 | cDNA cloning |
| MMP23B | MMP23B-CDS.R | CGCGGACCCCCCAGGAGTA | ||
| MMP23B | MIB2.3003 | GCTACCGGCAGATGGAGGAACGC | ||
| MMP23B | MMP23B.258 | GGTCAGCGTGTAGCGGCGTCTTC | ||
| MMP23B | 3'RACE.MMP23B.R | TACCCCGTCAACCACACCGACTGCC | ||
| MMP23B | 3'RACE nest | GACTTCTGCTACGAGTTCCC | ||
| MMP19 | 19.116 | GGCTTCCTACTCCCCATGAC | 60 | Intron 1Intron 2 |
| MMP19 | 19.336 | CTCATACGGACCCTTGTGGC | ||
| MMP19 | 19.311 | GATGATGCCACAAGGGTCCG | 58 | Intron 3 |
| MMP19 | 19.601 | GTTATTGGAGCAGTATGGGC | ||
| MMP19 | 19.501 | GTGGCTCCCTTGACTTTCCG | 56 | Intron 4 |
| MMP19 | 19.775 | CTGGGTGTATCGGGAGTG | ||
| MMP19 | 19.715 | CATCATTGCAGCCCACGAAC | 59 | Intron 5,6,7 |
| MMP19 | 19.1085 | CTTCCCAAAGGGCAGACAC | ||
| MMP19 | 19.1031 | ACCGTGACAGATTCAGGGC | 59 | Intron 8 |
| MMP19 | 19.2114 | AATCCGAGGCTCAACGACG | ||
| MMP19 | 19map1 | TGCTCAACGGCACCCTAC | 58 | mapping |
| MMP19 | 19map2 | AGGAAGGCGGTCAGAGTC | ||
| MMP23B | 23map1 | CAAAGAGCGGCACCGAAAGC | 60 | |
| MMP23B | 23map2 | GTGGTCCCAACGTAGCCTG | ||
| MMP19 | MMP19V1-F | TCGGACACAATGGATTCACTTC | expression analyses | |
| MMP19 | MMP19V1-R | GCCCGGAGACATCTTGAAATT | ||
| MMP19 | MMP19V1-P | FAM-CGTAGCGCCACACCTTGTCTCCCT-TAMRA | ||
| MMP23B | MMP23B-F | TCGCAGGACGAGCTGTGG | ||
| MMP23B | MMP23B-R | GCGTCACAGAAGCCCCTC | ||
| MMP23B | MMP23B-P | FAM-CACACGAACAGCCGGTCCAGG-TAMRA | ||
| β-actin | ATCB-F | TCCAGAGGCGCTCTTCCA | ||
| β-actin | ATCB-R | CGCACTTCATGATCGAGTTGA | ||
| β-actin | ATCB-P | FAM-CCTCCTTCCTGGGCATGGAGTCCT-TAMRA | ||
| MMP19 | 19mor3023:exon 5 | ACTCTGACCGCCTTCCTC | 60 | SNP genotyping |
| MMP19 | 19mor3367:exon 5 | TCAAGCCCGAAGAGGTGC | ||
| MMP23B | 23mor2.1:exon 3 | AGAGCGGCACCGAAAGCG | 60 | |
| MMP23B | 23mor2.2:exon 3 | CTGCTTGACCCCAGACCC | ||
| MMP23B | 23mor260:exon 4 | CGGCGGCATCCACTTTGACG | 60 | |
| MMP23B | 23mor2.2:exon 4 | CTGCTTGACCCCAGACCC |
Mapping of porcine MMP19 and MMP23B genes
The chromosome assignment of porcine MMP19 and MMP23B was performed with the INRA-University of Minnesota 7000 porcine radiation hybrid panel (IMpRH), consisting of 118 hamster porcine hybrid cell lines 21. PCR reactions for IMpRH mapping were carried out in a volume of 20 µL containing 100 ng DNA, 2 µL 10x PCR buffer, 1.6 µL 2.5 mM dNTP mix, 0.5 µL sense primer (100 ng/µL), 0.5 µL anti-sense primer (100 ng/µL), 0.2 µL 5 U LA Taq DNA polymerase (Fermentas, Vilnius, Lithuania) and sterile water. The PCR amplification was initiated with a 5-minute denaturation at 95°C, followed by 35 cycles of 95°C for 30 sec, Tm for 30 sec and 72°C for 30 sec, and a final extension 72°C for 4 min. PCR products were analyzed by 2% agarose gel electrophoresis and each clone in the IMpRH panel was amplified three repeats to get reliable results. Typing results were recorded and submitted to https://www-lgc.toulouse.inra.fr/pig/RH/IMpRH.htm for analysis. The primer pairs used for IMpRH mapping of porcine MMP19 and MMP23B genes are shown in Table 1.
Expression patterns of porcine MMP19 and MMP23B genes
Quantitative real-time PCR was employed to detect temporal and spatial expression of porcine MMP19 and MMP23B genes. Total RNA was extracted from tissue samples using Trizol Reagent (Invitrogen, Carlsbad, CA, USA) and cDNA was prepared using the reverse transcription kit (Takara, Dalian, China) according to the manufacturer's procedures. Each 20µL real-time PCR reaction contained 100 ng cDNA, 2.0 µL 10x PCR buffer, 0.4 µL 10 mM dNTP, 3.6 µL 5 μM sense primer, 1.2 µL 5 μM antisense primer, 0.2 µL 5 U/µL r-Taq DNA polymerase, 0.6 µL 5 μM probe, 0.5 µL DMSO, 4 µL 10 M BETAINE and sterile water. The PCR reaction was performed in triplicate and carried out at 95°C for 5 min, followed by 40 cycles of 15 sec at 95°C and 1 min at 60°C on a 7500 Real-Time PCR System (Applied Biosystems, USA). The gene expression levels were quantified relative to the expression of GAPDH using Gene Expression Macro software (BIO-RAD, USA) and the results were analyzed by the 2-ΔΔCt method 22. The primer pairs and hydrolysis probes for expression analysis are listed in Table 1.
SNP detection and association analysis
Primer pairs were designed for SNP scanning of our porcine MMP19 and MMP23B genes, and SNP sites were identified by sequencing PCR products from 4 pig breeds: Yorkshire, Landrace, Tongcheng and Wuzhishan pigs. Consequently, three SNPs C203T in exon 5 of MMP19 (MseI site), C131T in exon 3 (MboІ site) and A150G in exon 4 of MMP23B (Bsh1236I site) were discovered.
The PCR 20 µL reaction consisted of 20 ng DNA template, 2 µL 10x PCR buffer, 1.6 µL 2.5 mM dNTP mix, 0.5 µL sense primer (100 ng/µL), 0.5 µL antisense primer (100 ng/µL), 0.2 µL 5 U r-Taq DNA polymerase (Fermentas, Vilnius, Lithuania) and sterile water. The PCR reaction comprised of the initial denaturation at 95 °C for 5 min, 33 cycles of 30 sec at 94 °C, 30 sec at 60°C, and 30 sec at 72°C, followed by a final extension at 72 °C for 5 min. The RFLP reaction mixture consisted of 1 μL 10x buffer, 10 U restriction enzyme (NEB, Ipswich, MA, USA), 5 μL PCR products and sterile water. Samples were incubated at the specific temperature for restriction enzyme overnight. The digested products were analyzed by 2% agarose gel electrophoresis, stained with ethidium bromide, and visualized under UV illumination. The primer pairs for SNP genotyping are displayed in Table 1.
The association analysis was performed using the GLM procedure (SAS, 2006). Yijk =μ+ Bi + Gj +εijk 20, Where Yijk is the phenotypic value of a target trait; μ represents the population mean, Bi is the combination effect, Gj is the genotype effect and εijk represents the random error.
Results and Discussion
Molecular cloning and sequence analysis of porcine MMP19 and MMP23B genes
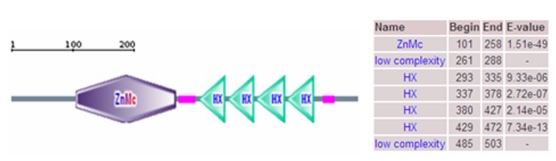
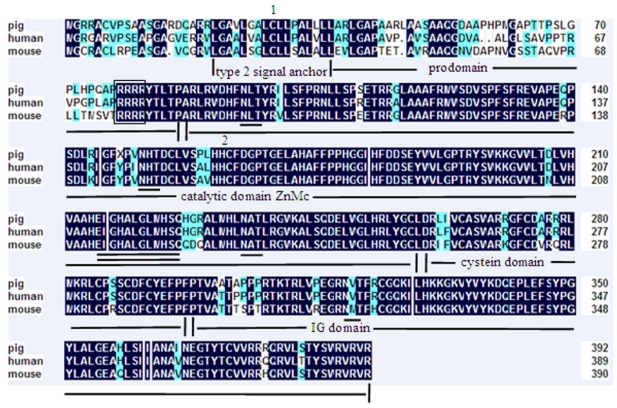
A 2258 bp cDNA contig of porcine MMP19 gene was assembled after sequencing the PCR and RACE products. This contig contains an ORF of 1623 bp, flanked by an 87 bp 5'UTR and a 548 bp 3'UTR. A consensus AATAAA polyadenylation signal was identified at 21 bp upstream of the poly(A) stretch. The porcine MMP19 was predicted to encode a protein of 540 amino acid residues (http://genes.mit.edu/GENSCAN.html) with a calculated molecular mass of 61.03218 kDa and an isoelectric point of 6.13 (http://weblab.cbi.pku.edu.cn/program.inputForm.do?program=pepstats(v6.0.1)). The deduced amino acid sequence shares 84% and 76% sequence similarity with human (NP_002420.1) and mouse (NP_067387.1) homologues, respectively. However, the porcine MMP19 has additional 32 and 13 amino acids more than human and mouse, respectively (shown in Fig. 1). Amino acid sequence analysis of pig MMP19 revealed that the precursor protein contains zinc-dependent metalloprotease (ZnMc) and hemopexin-like repeats (HX), similar to the domains of the human (Q99542) and mouse (Q9JHI0) MMP19 proteins (shown in Fig. 1 and Fig. 3).
Fig 1.
Amino acid sequence alignment of porcine, human and mouse MMP19. Note: Shading shows identical amino acid residues among the three species. Common structural domains are underlined. The MMPs consensus zinc-binding motif, HEXGHXXGXXH, is double underlined. Potential N-linked glycosylation sites are underlined. The conserved pro-domain cysteine residue is marked by the number 1 and the methionine residue is marked by the number 2.
Fig 3.
Domain of porcine MMP19 protein. Note: The porcine MMP19 precursor protein contains zinc-dependent metalloprotease (ZnMc) and hemopexin-like repeats (HX).
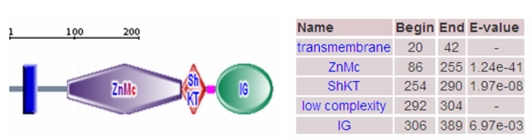
For the porcine MMP23B gene, a 1254 bp cDNA contig was assembled after sequencing the PCR and RACE products. This contig contains a 69 bp 3'UTR and an 1182 bp ORF encoding a protein of 393 amino acids. The predicted protein has a calculated molecular mass of 44.06404 kDa and an ioselectic point of 10.052. The deduced amino acid sequence shares 91%, 83%, 82%, 89%, 93% and 67% sequence identity with bovine (NP_001033645.1), rat (NP_446058.1), mouse (NP_036115), human (NP_008914.1), dog (XP_848890.1) and chicken (XP_417569.1) homologs, respectively (shown in Fig. 3 and Fig. 5). The amino acid sequence analysis showed that the porcine MMP23B precursor protein contains a transmembrane segments, zinc-dependent metalloprotease (ZnMc), ShK toxin domain (ShKT) and immunoglobulin (IG), similar to the domains of the human (O75900) and mouse (O88676) MMP23 proteins (shown in Fig. 3 and Fig. 4). The MMP23 ShKT domains from humans to hydra exhibit remarkable sequence conservation 23, indicating that it contributes critical role throughout the plant and animal kingdoms. Online analysis of the approximate 2000-bp fragment upstream of the pig MMP23B gene exon 1 indicated that this sequence contains no TATA box or CAAT box, but a classical CpG island promoter. This CpG island is 506bp in length, with 70.75% GC content and is rich in transcription factors binding sites including Sp1+, Myod+, Snail+, MZF1-, c-Ets+ and c-FOS (shown in Fig. 6).
Fig 5.
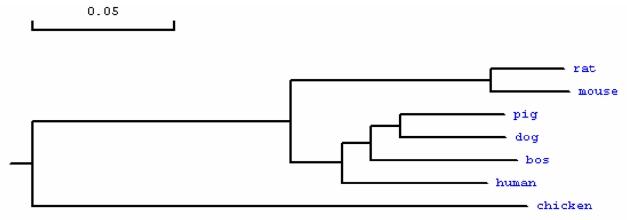
Phylogenetic tree of MMP23B in different species. Note: The GenBank accession numbers of those sequences are as follows: human: NP_008914.1, bos: NP_001033645.1, rat: NP_446058.1, mouse: NP_036115, dog: XP_848890.1, chicken: XP_417569.1. The porcine MMP23B amino acid sequence was deduced from the full cDNA sequence in this study.
Fig 4.
Domain of porcine MMP23B protein. Note: The porcine MMP23B precursor protein contains a transmembrane segments, zinc-dependent metalloprotease (ZnMc), ShK toxin domain (ShKT) and immunoglobulin (IG).
Fig 6.
Structure of pig MMP23B promoter. Note: The CpG island of the pig MMP23B promoter is shaded. The binding sites of the transcription factor are framed, and the corresponding transcription factors are shown beneath the binding sites. The transcription start site is indicated by triangles ▼. The start codon of the pig MMP23B gene is underlined. The symbol + represents transcription factor binding on the sense strand. The symbol - refers to the transcription factor binding on the antisense strand.
The genomic sequence of the porcine MMP19 gene is 6369 bp containing 9 exons and 8 introns, while the MMP23B gene is 2343 bp with 8 exons and 7 introns. Based on the analysis of the genomic structure, the exon/intron junction of MMP19 and MMP23B all conform to the GT-AG rule (shown in Table 2).
Table 2.
Structure of pig MMP19 and MMP23B genes
| Gene | Number | Exon size(bp) | Intron size (bp) | 5' splice donor | 3' splice acceptor |
|---|---|---|---|---|---|
| MMP19 | 1 | 177 | 283 | GTG/GTGAG | TCATAG/GATTAT |
| 2 | 86 | 1467 | TGAG/GTGAGA | TTTTCAG/AGCTT | |
| 3 | 131 | 244 | GCTGG/GTGA | ACCAG/GCC | |
| 4 | 216 | 451 | CTG/GTAGGT | TCCAG/GGAGGG | |
| 5 | 246 | 444 | CTATG/GTCAG | TGCAG/GCAAG | |
| 6 | 126 | 684 | TGG/GTAAGAC | CTCAG/GGCC | |
| 7 | 165 | 350 | TTAAGG/GTAAC | CTCTCAG/GAG | |
| 8 | 128 | 188 | TAAG/GTACA | TACCTAG/GGCT | |
| 9 | 953 | ||||
| MMP23B | 1 | 162 | 336 | CAA/GTGAG | GAAG/GG |
| 2 | 137 | 79 | CAG/GTGCG | CCCAG/GATCC | |
| 3 | 140 | 77 | ATAG/GTGGG | CCTAG/GCTTCT | |
| 4 | 168 | 368 | AG/GTGAC | CGCAAG/GCGT | |
| 5 | 165 | 77 | ACG/GTGAGT | TCCCGCAG/GCT | |
| 6 | 111 | 78 | TACG/GTGAG | AGCAG/AGTTC | |
| 7 | 127 | 78 | AGTGTA/GTGA | CCCGCAG/CTGG | |
| 8 | 216 |
Chromosome assignment of porcine MMP19 and MMP23B genes
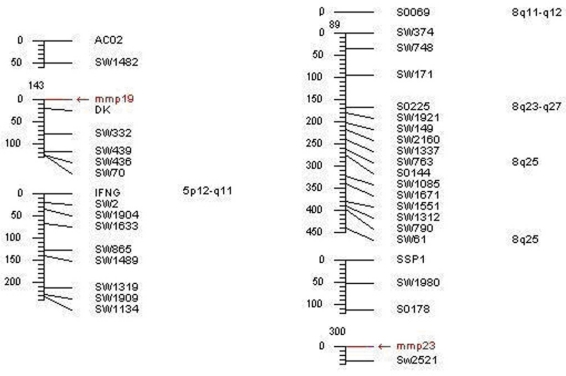
The data analysis of the somatic cell hybrid panel revealed that porcine MMP19 was mapped to the region of swine chromosome 5p12-q11 (LOD score=18.02) and closely linked to marker DK, the MMP23B gene was mapped to 8q11-q12 (LOD score=11.69) and tightly linked to the microsatellite marker Sw2521 (shown in Table 3 and Fig. 7). The mapping information of these genes is in consistent with our isolated porcine genome sequence. In addition, the MMP19 gene was assigned to chromosome 12q14 in human and chromosome 10 D3 in mouse, while MMP23B was assigned to chromosome 1p36.3 in human and chromosome 4 E2 in mouse. Our results agree with the comparative mapping data, as human chromosome 12q14 is a syntenic region of porcine chromosome 5 24. Therefore, our result gives a precise location of porcine MMP19 and MMP23B and enriches the comparative map information between HSA 12 and SSC 5.
Table 3.
RH mapping results of MMP19 and MMP23B genes
| Genes | Cytogenetic position | Linked markers | Retention | Breakage frequency | RH Dist (Ray) |
LOD score |
|---|---|---|---|---|---|---|
| MMP19 | SSC5 | DK | 44 | 0.19 | 0.21 | 18.02 |
| MMP23B | SSC8 | Sw2521 | 26 | 0.28 | 0.33 | 11.69 |
Fig 7.
Mapping results of porcine MMP19 and MMP23B genes.Note: The left refers to the porcine MMP19 gene location, and the right represents the pig MMP23B gene mapping result.
Temporal and spatial expression of porcine MMP19 and MMP23B gene
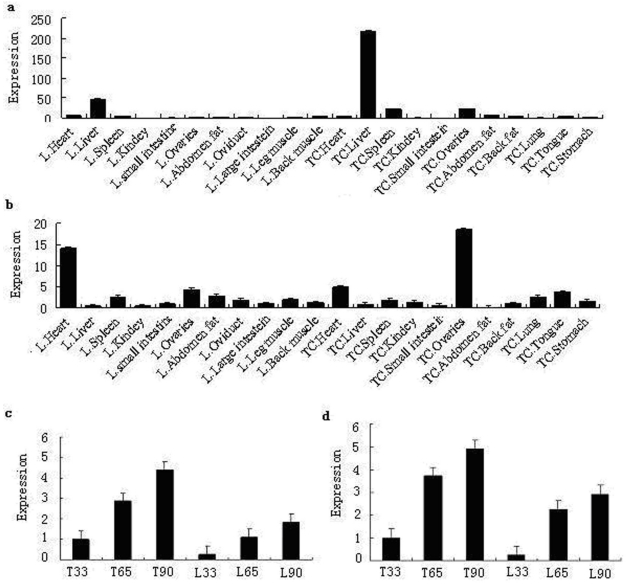
Quantitative real-time PCR analysis showed that while the porcine MMP19 gene was highly expressed in the liver of both breeds, it is much higher in Tongcheng pigs than in Landrace pigs. In both breeds, MMP19 was weakly expressed in other tissues. Porcine MMP23B was expressed at high levels in the heart and ovarian of both Landrace and Tongcheng pigs. Otherwise, it was expressed ubiquitously at low levels in both breeds. In ovarian tissue, however, the expression level of MMP23B in the Tongcheng pig was much higher than in the Landrace pig, while this expression pattern is reversed in the heart tissue. However, the expressions of porcine MMP19 and MMP23B are partial consistency with those of the human MMP19 and MMP23B 2, 12, respectively. These may be attributed to the distribution characteristics and biological function of these genes in different species.
The tissue expression patterns correspond with our mapping results that the porcine MMP23B gene was mapped to the Sus scrofa chromosome 8, where it is strongly linked to the microsatellite marker SW252. Within this region, several QTLs associated with reproduction traits, such as ovulation rate, teat number, as well as the FSH content in blood, are found. Moreover, previous reports indicated that MMPs play key roles in follicle rupture and they are up-regulated and activated in coordination with ovarian follicle rupture before ovulation 25.
By using RT-PCR, the temporal expression patterns of porcine MMP19 and MMP23B were examined in Tongcheng and Landrace pigs. As shown in Fig. 8c and Fig. 8d, porcine MMP19 and MMP23B genes were expressed at three stages and had the similar trend of expression: increasing during the development stages. The expression patterns indicated that MMP19 and MMP23B could play roles in porcine prenatal skeletal muscle development, the result is consistency with the MMPs functions of development and normal physiology 26. Moreover, the myoblast proliferation is an important molecular event in development of porcine skeletal muscle. This suggested that porcine MMP19 and MMP23B are involved in cell proliferation in myogenesis. Notably, the expression level in Tongcheng pigs is significantly higher than that in Landrace pigs. According to our previous study, Tongcheng pigs have a slower muscle growth rate than Landrace pigs 27, and the difference expression level of MMP19 and MMP23B may be associated with the differences between Tongcheng and Landrace pigs in the intensity and timing of myoblast fusion during prenatal myogenesis.
Fig 8.
Expression pattern analysis by real-time PCR method. Note: a and b are the tissue expression results of porcine MMP19 and MMP23B in Tongcheng and Landrace pigs, respectively. c and d are the period expression patterns during prenatal skeletal muscle development in Tongcheng and Landrace pigs. L and TC refer to Landrace and Tongcheng pig for porcine MMP19 and MMP23B genes, respectively. 33, 65 and 90 indicate days post coitus (dpc), respectively.
SNP detection, allele frequency and association analysis
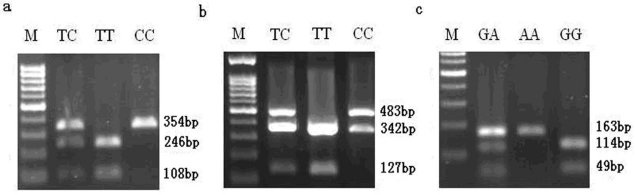
Three SNP sites were discovered through scanning DNA sequence of porcine MMP19 and MMP23B genes. The three SNPs, C203T in exon 5 with an MseІ site in MMP19, C131T in exon 3 with an MboІ site and A150G in exon 4 with a Bsh1236I site in MMP23B, were genotyped by PCR-RFLP technique in the more than 200 pigs in our experimental population. After digestion by MseI, the 354 bp PCR products of MMP19 were digested into 246 bp and 108 bp fragments (allele T) (Fig. 9a). In addition to the MboI site at 131 bp in exon 3 of MMP23B, the fragment used for genotyping contains another MboI site, and after digestion, the 825 bp PCR amplicon produced three fragments of 342 bp, 356 bp and 127 bp for allele T or two fragments of 483 bp and 342 bp for allele C. However, the 342 bp and 356 bp fragments were not distinguished in our study (Fig. 9b). For exon 4 of MMP23B, the 163 bp PCR amplicon can be digested by Bsh1236I, producing 114 bp and 49 bp fragments for allele G (Fig. 9c).
Fig 9.
SNP genotyping results by PCR-RFLP method. Note: a is the PCR-MseI-RFLP analysis result of SNP at C203T in exon 5 of the MMP19 gene; b is the PCR-MboI-RFLP analysis at C131T in exon 3 of the MMP23B gene; c is the PCR-Bsh1236I-RFLP analysis at A150G in exon 4 of the MMP23B gene. The genotypes are shown on the top lanes; M refers to the DNA molecular weight marker.
Association analysis results indicated that C203T in exon 5 of MMP19 has a significant association with the blood parameters WBC (g/L) and IgG2 (mg/mL) (P<0.05). Furthermore, the WBC in the genotype CT individuals (32.72 ± 2.986) is significantly more than in the genotype TT individuals (19.67 ± 5.23) (P<0.05). Genotype CT individuals (55.06 ± 2.287) have significantly higher levels of IgG2 than the genotype TT (45.81 ± 3.99) and CC (46.919 ± 2.85) individuals (P<0.05). Consistent with our mapping results, the porcine MMP19 gene was mapped to the Sus scrofa chromosome 5 and is strongly linked to the marker DK; in this region, a QTL exists for IgG2 of K88 E. coli. Moreover, the results also agree with the immunity function of MMP19 gene involving breast cancer, nasopharyngeal carcinoma, brain, lung, ovarian cancer and so on 28-30. MMP23B, C131T in exon 3 is associated with the blood parameters HGB (g/L) and MCH significantly (P<0.05). SNP A150G in exon 4 has no significant association with the economic traits in pigs.
Overall, the temporal expression patterns and the association results of MMP19 and MMP23B indicated that the genes may play roles in porcine immunity and embryonic development. The results correspond with the MMPs functions involving regulate both development and immunity in the Tribolium Model Insect 31.
In conclusion, we cloned and characterized the porcine MMP19 and MMP23B genes. Chromosome localization by the INRA IMpRH panel assigned the porcine MMP19 gene to SSC5p12-q11, closely linked to marker DK, and the MMP23B gene to SSC8q11-q12, tightly linked to microsatellite marker Sw2521. We have also analyzed their tissue expression differences and skeletal muscle development stages by real-time PCR in Landrace and Tongcheng pigs. We provided initial evidence that porcine MMP19 and MMP23B genes were ubiquitously expressed in tissues and similarly expressed in prenatal skeletal muscle development. Genotyping and association analyses were performed on the SNPs MseI, MboI and Bsh1236I and revealed that MMP19 has a significant association with the blood parameters WBC (g/L) (P<0.05) and IgG2 (mg/mL) (P<0.05), MMP23B was significantly associated with the blood parameters HGB (g/L) (P<0.05) and MCH (P<0.05). Our information provides an important molecular basis for further studies on the functions of porcine MMPs genes and the identified polymorphisms that may serve as genetic markers for immune traits in animal breeding.
Fig 2.
Amino acid sequence alignment of porcine, human and mouse MMP23B. Note: Shading shows identical amino acid residues among the three species. Common structural domains are underlined. The MMPs consensus zinc-binding motif, HEXGHXXGXXH, is double underlined. Potential N-linked glycosylation sites are underlined. The Flynn cleavage site is framed. The conserved pro-domain cystein residue is marked by the number 1 and the methionine residue is marked by the number 2.
Table 4.
Allele frequency in different pig breeds
| SNPs of MMP19 | Pig breeds | ||||||||||
| Tongcheng pig | Yorkshire pig | Landrace pig | LYT pig | YLT pig | |||||||
| C203T in exon 5 | |||||||||||
| NO. of CC individuals | 2 | 12 | 3 | 8 | 18 | ||||||
| NO. of CT individuals | 13 | 1 | 6 | 23 | 10 | ||||||
| NO. of TT individuals | 14 | 0 | 0 | 6 | 0 | ||||||
| Frequency of C | 0.29 | 0.96 | 0.57 | 0.53 | 0.82 | ||||||
| Frequency of T | 0.71 | 0.04 | 0.33 | 0.47 | 0.18 | ||||||
| SNPs of MMP23B | Pig breeds | ||||||||||
| Tongcheng pig | Yorkshire pig | Landrace pig | Wuzhishan pig | Bama pig | Laiwu pig | Guizhou pig | |||||
| C131T in exon 3 | |||||||||||
| NO. of CC individuals | 4 | 12 | 0 | 6 | 0 | 0 | 7 | ||||
| NO. of CT individuals | 13 | 3 | 8 | 25 | 7 | 11 | 20 | ||||
| NO. of TT individuals | 18 | 0 | 3 | 8 | 25 | 31 | 15 | ||||
| Frequency of C | 0.30 | 0.90 | 0.37 | 0.47 | 0.11 | 0.87 | 0.41 | ||||
| Frequency of T | 0.70 | 0.10 | 0.63 | 0.53 | 0.89 | 0.13 | 0.59 | ||||
| A150G in exon 4 | |||||||||||
| NO. of GG individuals | 16 | 0 | 2 | 0 | 33 | 32 | 15 | ||||
| NO. of AG individuals | 12 | 5 | 9 | 39 | 11 | 10 | 20 | ||||
| NO. of AA individuals | 5 | 13 | 0 | 0 | 0 | 0 | 6 | ||||
| Frequency of G | 0.66 | 0.14 | 0.59 | 0.50 | 0.88 | 0.12 | 0.61 | ||||
| Frequency of A | 0.34 | 0.86 | 0.41 | 0.50 | 0.12 | 0.88 | 0.39 | ||||
Table 5.
Association analysis of different genotypes with traits
| traits | genotypes/number | P value | |||||
|---|---|---|---|---|---|---|---|
| C203T in exon 5 of MMP19 | |||||||
| WBC | 27.71±3.275 | 32.72±2.986 | 19.67±5.23 | 0.0472* | 0.278 | 0.2236 | 0.0183* |
| IgG2 | 46.919±2.85 | 55.06±2.287 | 45.81±3.99 | 0.0195* | 0.0459* | 0.8368 | 0.0256* |
| C131T in exon 3 of MMP23B | |||||||
| CC/27 | TC/77 | TT/35 | |||||
| HGB | 58.263±9.492 | 54.322±5.73 | 29.298±8.322 | 0.0189* | 0.7371 | 0.0326 | 0.0074** |
| MCH | 152.94±25.71 | 147.33±15.52 | 82.86±22.54 | 0.0304* | 0.8598 | 0.0558 | 0.0108* |
Acknowledgments
We are grateful to Dr. Martine Yerle for providing the IMpRH panel. We are also very grateful to Sanping Xu in the Husbandry Bureau of Tongcheng County for help in animal material collection. This research was supported by the National Key Project (2009ZX08010-022B), the National Natural Science Foundation of China (30830080), National Key Scientific Program (2009CB011604) and the Project of Science and Technology Innovation Team “Research and Improvement of Domestic Animal Germplasm”, Institute of Animal Science, Chinese Academy of Agricultural Science (ywf-td-1).
References
- 1.Sedlacek R, Mauch S, Kolb B. et al. Matrix metalloproteinase MMP-19 (RASI-1) is expressed on the surface of activated peripheral blood mononuclear cells and is detected as an autoantigen in rheumatoid arthritis. Immunobiology. 1998;198(4):408–423. doi: 10.1016/S0171-2985(98)80049-1. [DOI] [PubMed] [Google Scholar]
- 2.Pendas AM, Knauper V, Puente XS. et al. Identification and characterization of a novel human matrix metalloproteinase with unique structural characteristics, chromosomal location, and tissue distribution. J Biol Chem. 1997;272:4281–4286. doi: 10.1074/jbc.272.7.4281. [DOI] [PubMed] [Google Scholar]
- 3.Mysliwy J, Dingley AJ, Sedlacek R. et al. Structural characterization and binding properties of the hemopexin-like domain of the matrixmetalloproteinase-19. Protein Expr Purif. 2006;46:406–13. doi: 10.1016/j.pep.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 4.Hagglund AC, Ny A, Leonardsson G. et al. Regulation and localization of matrix metalloproteinases and tissue inhibitors of metalloproteinases in the mouse ovarian during gonadotropin-induced ovulation. Endocrinology. 1999;140:4351–4358. doi: 10.1210/endo.140.9.7002. [DOI] [PubMed] [Google Scholar]
- 5.Sadowski T, Dietrich S, Koschinsky F. et al. Matrix metalloproteinase 19 regulates insulin-like growth factor-mediated proliferation, migration, and adhesion in human keratinocytes through proteolysis of insulin-like growth factor binding protein-3. Mol Biol Cell. 2003;14:4569–4580. doi: 10.1091/mbc.E03-01-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jo M, Curry TEJr. Regulation of matrix metalloproteinase-19 messenger RNA expression in the rat ovarian. Biol Reprod. 2004;71:1796–1806. doi: 10.1095/biolreprod.104.031823. [DOI] [PubMed] [Google Scholar]
- 7.Lettau I, Hattermann K, Held-Feindt J. et al. Matrix Metalloproteinase-19 is Highly Expressed in Astroglial Tumors and Promotes Invasion of Glioma Cells. Journal of Neuropathology & Experimental Neurology. 2010;69:215–223. doi: 10.1097/NEN.0b013e3181ce9f67. [DOI] [PubMed] [Google Scholar]
- 8.Fox BP, Kandpal RP. EphB6 receptor significantly alters invasiveness and other phenotypic characteristics of human breast carcinoma cells. Oncogene. 2009;28:1706–1713. doi: 10.1038/onc.2009.18. [DOI] [PubMed] [Google Scholar]
- 9.Sadowski T, Dietrich S, Koschinsky F. et al. Matrix metalloproteinase 19 processes the laminin 5 gamma 2 chain and induces epithelial cell migration. Cell Mol Life Sci. 2005;62:870–880. doi: 10.1007/s00018-005-4478-8. [DOI] [PubMed] [Google Scholar]
- 10.Pendás AM, Folgueras AR, Llano E. et al. Diet-Induced Obesity and Reduced Skin Cancer Susceptibility in Matrix Metalloproteinase 19-Deficient Mice. Mol Cell Biol. 2004;24:5304– 5313. doi: 10.1128/MCB.24.12.5304-5313.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beck IM, Rückert R, Brandt K. et al. MMP19 Is Essential for T Cell Development and T Cell-Mediated Cutaneous Immune Responses. PLoS ONE. 2008;3(6):e2343. doi: 10.1371/journal.pone.0002343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Velasco G, Pendas AM, Fueyo A. et al. Cloning and characterization of human MMP-23, a new matrix metalloproteinase predominantly expressed in reproductive tissues and lacking conserved domains in other family members. J Biol Chem. 1999;274:4570–4576. doi: 10.1074/jbc.274.8.4570. [DOI] [PubMed] [Google Scholar]
- 13.Ohnishi J, Ohnishi E, Jin M. et al. Cloning and characterization of a rat ortholog of MMP-23 (matrix metalloproteinase-23), a unique type of membrane-anchored matrix metalloproteinase and conditioned switching of its expression during the ovarian follicular development. Mol Endocrinol. 2001;15:747–764. doi: 10.1210/mend.15.5.0638. [DOI] [PubMed] [Google Scholar]
- 14.Clancy BM, Johnson JD, Lambert A-J. et al. A gene expression profile for endochondral bone formation: oligonucleotide microarrays establish novel connections between known genes and BMP-2-induced bone formation in mouse quadriceps. Bone. 2003;33:46–63. doi: 10.1016/s8756-3282(03)00116-9. [DOI] [PubMed] [Google Scholar]
- 15.Davidson RK, Waters JG, Kevorkian L. et al. Expression profiling of metalloproteinases and their inhibitors in synovium and cartilage. Arthritis Res Ther. 2006;8:R124. doi: 10.1186/ar2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gajecka M, Yu W, Ballif BC. et al. Delineation of mechanisms and regions of dosage imbalance in complex rearrangements of 1p36 leads to a putative gene for regulation of cranial suture closure. Eur J Hum Genet. 2005;13:139–149. doi: 10.1038/sj.ejhg.5201302. [DOI] [PubMed] [Google Scholar]
- 17.Fortunato SJ, Menon R. Screening of novel matrix metalloproteinases (MMPs) in human fetal membranes. J Assist Reprod Genet. 2002;19:483–486. doi: 10.1023/A:1020362519981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J, Deng X, Demetriou AA. et al. Factors released from cholestatic rat livers possibly involved in inducing bone marrow hepatic stem cell priming. Stem Cells Dev. 2008;17:143–155. doi: 10.1089/scd.2007.0094. [DOI] [PubMed] [Google Scholar]
- 19.Tang ZhL, Zhang XJ, Yang ShL. et al. The chromosomal localization, expression pattern and polymorphism analysis of porcine FSCN1 gene differently expressed from LongSAGE library. Mol Biol Rep. 2010;37(5):2361–2367. doi: 10.1007/s11033-009-9742-9. [DOI] [PubMed] [Google Scholar]
- 20.Wang HL, Wang H, Zhu ZhM. et al. Subcellular localization, expression patterns, SNPs and association analyses of the porcine HUMMLC2B gene. Mol Gen Genomics. 2006;276:264–272. doi: 10.1007/s00438-006-0142-8. [DOI] [PubMed] [Google Scholar]
- 21.Yerle M, Pinton P, Robic A. et al. Construction of a whole-genome radiation hybrid panel for high-resolution gene mapping in pigs. Cytogenet Cell Genet. 1998;82:182–188. doi: 10.1159/000015095. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods. 2001;25:402–428. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Rangaraju S, Khoo KK, Feng ZP. et al. Potassium channel modulation by a toxin domain in matrix metalloprotease 23. J Biol Chem. 2010;285(12):9124–9136. doi: 10.1074/jbc.M109.071266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goureau A, Yerle M, Schmitz A. et al. Human and porcine correspondence of chromosome segments using bidirectional chromosome painting. Genomics. 1996;36:252–262. doi: 10.1006/geno.1996.0460. [DOI] [PubMed] [Google Scholar]
- 25.Curry TEJr, Osteen KG. Cyclic Changes in the Matrix Metalloproteinase System in the Ovarian and Uterus. Biol Reprod. 2001;64(5):1285–1296. doi: 10.1095/biolreprod64.5.1285. [DOI] [PubMed] [Google Scholar]
- 26.Thiennu H. Vu and Zena Werb. Matrix metalloproteinases: effectors of development and normal physiology. Genes Dev. 2000;14:2123–2133. doi: 10.1101/gad.815400. [DOI] [PubMed] [Google Scholar]
- 27.Tang ZhL, Li Y, Wan P. et al. LongSAGE analysis of skeletal muscle at three prenatal stages in Tongcheng and Landrace pigs. Genome Biology. 2007;8:R115. doi: 10.1186/gb-2007-8-6-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan KC, Ko JM, Lung HL. et al. Catalytic activity of matrix metalloproteinase-19 is essential for tumor suppressor and anti-angiogenic activities in nasopharyngeal carcinoma. Int J Cancer. 2011 doi: 10.1002/ijc.25855. doi: 10.1002/ijc.25855. [DOI] [PubMed] [Google Scholar]
- 29.Delassus GS, Cho H, Eliceiri GL. New signaling pathways from cancer progression modulators to mRNA expression of matrix metalloproteinases in breast cancer cells. J Cell Physiol. 2011 doi: 10.1002/jcp.22694. doi: 10.1002/jcp.22694. [DOI] [PubMed] [Google Scholar]
- 30.Delassus GS, Cho H, Hoang S. et al. Many new down- and up-regulatory signaling pathways, from known cancer progression suppressors to matrix metalloproteinases, differ widely in cells of various cancer. J Cell Physiol. 2010;224(2):549–58. doi: 10.1002/jcp.22157. [DOI] [PubMed] [Google Scholar]
- 31.Knorr E, Schmidtberg H, Vilcinskas A. et al. MMPs Regulate both Development and Immunity in the Tribolium Model Insect. PLoS ONE. 2009;4(3):e4751. doi: 10.1371/journal.pone.0004751. [DOI] [PMC free article] [PubMed] [Google Scholar]