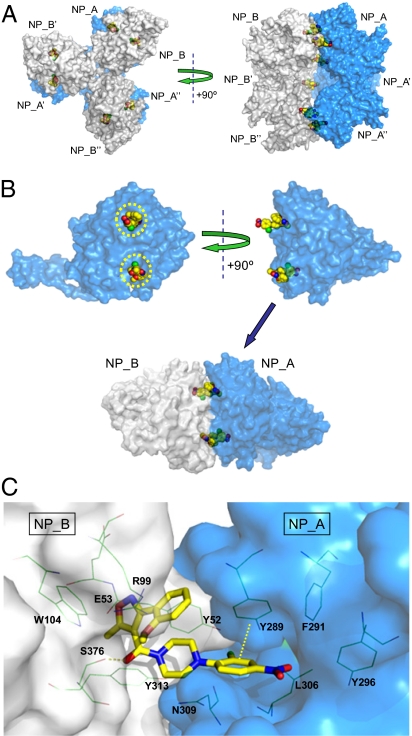
Fig. 4.
X-ray structure of compound 3:NP complex solved with a 2.7-Å resolution. (A) Six molecules of compound 3 (yellow) bridge two NP trimers (NP_A, NP_A′, and NP_A′′ in blue; NP_B, NP_B′, and NP_B′′ in gray) to form a dimer of trimers (or hexamer). (B) Two molecules of compound 3 (yellow) bridge an NP_A:NP_B dimeric subunit (NP_A in blue; NP_B in gray) in an antiparallel orientation. Yellow circles depict the regions of NP that were sensitive to amino acid substitution as described in Fig. 1. (C) Detailed view of compound 3 bound to an NP_A:NP_B dimeric subunit (NP_A in blue; NP_B in gray). Key ligand:protein contacts include a hydrogen bond from the amide carbonyl of compound 3 to the S376 side chain of NP_B and a face-to-face π–π interaction between the 2-Cl-4-NO2-Ph ring of compound 3 and Y289 of NP_A.