Abstract
Recent studies have revealed that 2–3% of the genome of non-Africans might come from Neanderthals, suggesting a more complex scenario of modern human evolution than previously anticipated. In this paper, we use a model of admixture during a spatial expansion to study the hybridization of Neanderthals with modern humans during their spread out of Africa. We find that observed low levels of Neanderthal ancestry in Eurasians are compatible with a very low rate of interbreeding (<2%), potentially attributable to a very strong avoidance of interspecific matings, a low fitness of hybrids, or both. These results suggesting the presence of very effective barriers to gene flow between the two species are robust to uncertainties about the exact demography of the Paleolithic populations, and they are also found to be compatible with the observed lack of mtDNA introgression. Our model additionally suggests that similarly low levels of introgression in Europe and Asia may result from distinct admixture events having occurred beyond the Middle East, after the split of Europeans and Asians. This hypothesis could be tested because it predicts that different components of Neanderthal ancestry should be present in Europeans and in Asians.
Keywords: genetic introgression, simulation
Recent analyses have revealed that Neanderthal genomes show closer genetic affinities with contemporary non-Africans than with West Africans (1, 2). Although this could be attributable to the existence of ancient subdivisions within Africa, it seems better explained by ancient episodes of admixture between Neanderthals and early Eurasians (1, 2). These results are at odds with those obtained from mtDNA (3–5) and challenge the simplest version of an out-of-Africa model of human evolution, which posits a complete replacement of Neanderthals by modern humans (e.g., 6, 7). It is thus likely that anatomically modern humans (Homo sapiens) have hybridized with Neanderthals (1), Denisovans (2), and potentially other archaic humans (e.g., 8, 9) while migrating out of Africa. However, the current quantification of the introgression does not shed much light on the hybridization process itself, which remains relatively unclear.
For instance, similar levels of Neanderthal introgression are observed in Europe and in Asia (1), which has been interpreted as evidence for a single and limited episode of admixture between Neanderthals and the ancestors of Eurasians some 50–60 kya (1). This interpretation implies either that there has been no subsequent admixture between modern humans and Neanderthals when the formers colonized Europe some 40 kya (10–12) or that some admixture occurred in Europe, where these species coexisted (13), but that this signal has now disappeared because of drift (14) or later migrations of nonadmixed populations (1). Moreover, the very low level of Neanderthal ancestry observed in Eurasians (1.9–3.1%) (2) is somehow surprising, because one would expect to see massive levels of Neanderthal introgression into modern humans if admixture was not strongly prevented during the range expansion of modern humans out of Africa (3, 15).
To examine these issues and clarify the process of hybridization between Neanderthals and modern humans, we have used a realistic and spatially explicit model of admixture and competition between modern humans and Neanderthals (3). Using extensive simulations, we have estimated the interbreeding success rate between humans and Neanderthals as well as the spatial scale of hybridization that is compatible with the observed patterns of Neanderthal ancestry in contemporary humans, assuming that the latter migrated out of Africa into Eurasia 50 kya (6, 7).
Results
Low Rates of Interbreeding Between Humans and Neanderthals.
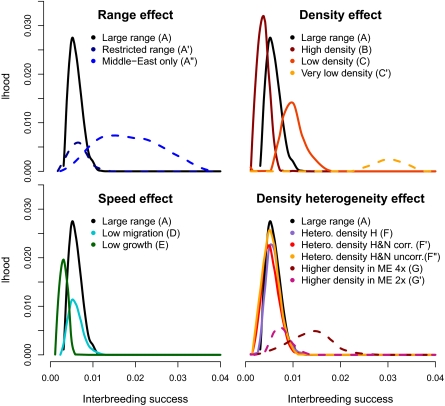
Using spatially explicit simulations, we have computed the expected amount of Neanderthal ancestry in present-day samples from Europe (France) and Asia (China) for different levels of admixture with Neanderthals and over various possible Neanderthal ranges (Fig. 1). Under our model of admixture during range expansion, we find that observed low levels of Neanderthal introgression into Eurasians imply the existence of extremely strong barriers to gene flow between the two species (Table 1) because of a very low fitness of human-Neanderthal hybrids, a very strong avoidance of interspecific mating, or a combination of these pre- and postzygotic barriers. Indeed, under most investigated demographic scenarios, the interbreeding success rate between humans and Neanderthals was found to be below 2% (Fig. 2, Table 1, Fig. S1, and Table S1). Under demographic scenario A (Table 1), which is based on the most realistic demographic parameters, we estimate this interbreeding success to be even well below 1% (0.51%, mode of black curve in Fig. 2; 95% confidence interval: 0.33–0.89%). Higher estimates of interbreeding success are obtained by assuming lower local population densities (scenarios C and C′), that hybridization between the two species only occurred in a small area of the Middle East (1.5%, scenario A′′; Fig. 2 and Table 1), or that population densities were higher in the Middle East than in other regions of Europe or Asia (scenarios G and G′). Contrastingly, lower interbreeding success estimates are obtained under demographic scenarios where the two species can interact for a longer time, for example, where local population densities are higher (scenario B, 0.37%) or where population growth is slower (scenario E, 0.30%). As can be seen in Fig. 2, interbreeding success generally decreases with increasing population densities and with decreasing expansion speeds, but it is relatively unaffected by heterogeneous densities over the Neanderthal and human ranges.
Fig. 1.
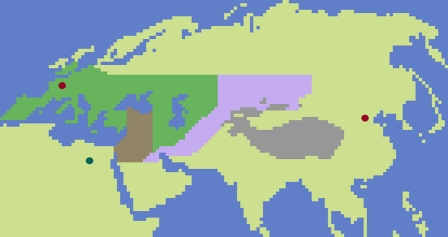
Simulated landscape used in our simulations. The union of the dark green and brown zones represents the conventionally assumed Neanderthal range (22) (scenario A′ in Table 1), whereas the violet zone represents a larger range, including the Altai Mountains, where Neanderthals remains have been identified recently. The brown zone represents an even more restricted area of potential hybridization in the Middle East (scenario A′′ in Table 1). The gray zone is the Himalayan range, where migrations have been disallowed. The dark green dot is an arbitrary place of origin for the expansion out of Africa, and the two red dots are the locations of the two samples where introgression is measured (Paris, France and Beijing, China). In our simulations, the continental areas have been divided into square cells (cell size = deme area = 100 × 100 km2) where a human population and a Neanderthal local population could potentially coexist, compete, and exchange migrants.
Table 1.
Demographic parameters and simulation results for the various scenarios presented in this study
| Models | KN* | KH† | r‡ | mN§ | mH¶ | Colonization time∥ | Estimated interbreeding success** | Model A relative probability†† |
| A: Large Neanderthal range | 200 | 800 | 0.8 | 0.1 | 0.2 | 220 | 0.0051 (0.0033–0.0089) | — |
| A′: Restricted Neanderthal range | 200 | 800 | 0.8 | 0.1 | 0.2 | 220 | 0.0065 (0.0023–0.0107) | 4.66 |
| A′′: Hybridization in ME only | 200 | 800 | 0.8 | 0.1 | 0.2 | 180‡‡ | 0.0153 (0.0048–0.0342) | 3.74 |
| B: Large K | 400 | 1,600 | 0.8 | 0.1 | 0.2 | 220 | 0.0037 (0.0013–0.0061) | 0.86 |
| C: Small K | 100 | 400 | 0.8 | 0.1 | 0.2 | 220 | 0.0097 (0.0059–0.0150) | 2.26 |
| C′: Very small K | 25 | 100 | 0.8 | 0.1 | 0.2 | 240 | 0.0302 (0.0239–0.0363) | 12.8 |
| D: Small m | 200 | 800 | 0.8 | 0.05 | 0.1 | 290 | 0.0051 (0.0029–0.0092) | 2.42 |
| E: Small r | 200 | 800 | 0.4 | 0.1 | 0.2 | 300 | 0.0030 (0.0013–0.0049) | 1.40 |
| F: Variable KH | 200 | 200–1,600 | 0.8 | 0.1 | 0.2 | 220 | 0.0054 (0.0033–0.0090) | 1.21 |
| F′: Variable KH and KN (correlated) | 50–400 | 200–1,600 | 0.8 | 0.1 | 0.2 | 220 | 0.0049 (0.0021–0.0087) | 1.22 |
| F′′: Variable KH and KN (uncorrelated) | 50–400 | 200–1,600 | 0.8 | 0.1 | 0.2 | 220 | 0.0051 (0.0019–0.0089) | 1.07 |
| G: K fourfold higher in ME | 200 (50) | 800 (200) | 0.8 | 0.1 | 0.2 | 220 | 0.0147 (0.0046–0.0218) | 5.62 |
| G′: K twofold higher in ME | 200 (100) | 800 (400) | 0.8 | 0.1 | 0.2 | 220 | 0.0072 (0.0052–0.0116) | 4.72 |
All scenarios were simulated by assuming a deme area of 100 × 100 km2. All scenarios except A′ and A′′ assume that hybridization could occur over the large Neanderthal range shown in Fig. 1. Demographic parameters for scenario A are judged the most plausible, given our review of the literature (Materials and Methods). ME, Middle East.
*Neanderthal carrying capacity.
†Human carrying capacity.
‡Intrinsic rate of growth.
§Migration rate between Neanderthal demes.
¶Migration rate between human demes.
∥Approximate time (in generations) for the colonization of Europe from the Middle East as estimated from the simulations. This statistic is determined by the growth and migration rates, and it also varies slightly with the hybridization rate.
**Maximum-likelihood estimates of interbreeding success between humans and Neanderthals are reported with limits of a 95% confidence interval within brackets.
††Probability of scenario A relative to the other scenarios computed from weighted AICs (Material and Methods).
‡‡Hybridization occurs for about 80 generations in the Middle East.
Fig. 2.
Distribution of the proportion of simulations (among 10,000) resulting in Neanderthal introgression levels compatible with observations (1.9%–3.1%) (2) in both French and Chinese samples. Each likelihood curve corresponds to a different demographic scenario described in Table 1 and Fig. 1. Results were obtained by assuming a deme area of 100 × 100 km2 (results obtained under a different map resolution are presented in Fig. S1). Solid lines correspond to scenarios that are equally likely (within 2 AIC units from the scenario with the highest likelihood), whereas scenarios shown with a dotted line have an associated AIC more than 2 units larger, and thus cannot be considered as equally well supported by the data. corr., correlated; lhood, likelihood; ME, Middle East; uncorr., uncorrelated.
We have identified with a dotted line in Fig. 2 all scenarios that are less supported by the data than the standard scenario A based on the Akaike Information Criterion (AIC) (16). The model's relative probabilities are also reported in Table 1. Based on the AIC, most alternative scenarios assuming a large Neanderthal range are equally well supported by the data, with the exception of scenarios with extreme parameter values (C′, G, and G′), which, interestingly, would all imply slightly larger interbreeding success.
Note that when we perform simulations by assuming that each Paleolithic subpopulation occupied a smaller territory (2,500 km2 instead of 10,000 km2), we obtain higher estimates of interbreeding success (Fig. S1 and Table S1), which, nevertheless, rarely exceed 2% for most scenarios. However, all these models have a lower relative probability than scenario A (Table S1).
Human-Neanderthal Hybridization Range.
Under our model of hybridization during range expansions, similar amounts of Neanderthal ancestry in France and China (Fig. S2) are more often observed if the geographical range of Neanderthals extended up to the Altai Mountains north of the Himalayas. Indeed, a hybridization range restricted to Europe, the Middle East, and the Caucasus region (including the brown and green areas in Fig. 1) would always lead to a much larger Neanderthal introgression level in Europe than in China (Figs. S2 and S3, light bars). Although we cannot define the exact geographical distribution of the Neanderthals, our results are thus in agreement with a Neanderthal Asian range extending to the Altai Mountains, where some hominin fossils of the Okladnikov site have been recently identified as Neanderthals on the basis of fossil DNA analyses (17, 18). It has to be noted that we obtain very similar estimates of interbreeding success by using either the large or the restricted Neanderthal range (0.51% vs. 0.65%, respectively; Fig. 2 and Table 1). Based on the AIC, the scenario with a large Neanderthal range is found to be 4.7 times more likely than that associated with a restricted range (scenario A′ in Fig. 1; ΔAIC = 3.1) and 3.7 times more likely than the model in which hybridization between the two species is only possible in a small area of the Near East (scenario A′′ in Fig. 1; ΔAIC = 2.63).
Discussion
The very low interbreeding success of human-Neanderthal hybrids that we obtain (<2%) is in keeping with previous analyses of mtDNA data (3, 4) that suggested very limited hybridization between Neanderthals and early modern humans (3, 4, 14). Note, however, that in our modeling, we cannot distinguish between prezygotic and postzygotic reproductive barriers (i.e., social, behavioral, ecological, genetic). It is only if mating was at random between humans and Neanderthals that this quantity could be considered as the fitness of the hybrids, but it is likely that assortative mating strongly contributed to this very low interbreeding success.
To check further if our estimate based on nuclear DNA introgression is compatible with the observed absence of Neanderthal mitochondrial lineages among contemporary humans (18), we ran additional simulations with deme densities adjusted to mtDNA (i.e., by using an effective deme density for mtDNA 4-fold smaller than for nuclear DNA). We thus simulated the genealogy of 20 samples of 100-mtDNA sequences scattered over all Eurasia, and we estimated the fraction of these current lineages to be of Neanderthal ancestry by using a conservative interbreeding success of 2% for the hybrids. Among 10,000 simulations of this process, we could never observe any mitochondrial sequence of Neanderthal origin in our samples. We thus conclude that an interbreeding success smaller than 2% for Neanderthal-human hybrids is fully compatible with limited Neanderthal nuclear introgression and with no introgression of mtDNA. Note that we obtained comparable estimates of maximum interbreeding success rates (<3%) in a previous simulation study that focused on the absence of Neanderthal mtDNA introgression into modern humans (figure 4 of ref. 3).
Under our model of range expansion of modern humans competing locally with Neanderthals, admixture can only occur in a narrow zone on the expansion front where the two populations coexist and have the possibility of hybridizing (3). To have similar final rates of introgression, the number of hybridizations having occurred on the Asian wave front must be roughly similar to that the number of hybridizations having occurred on the European wave front, which explains why the restricted Neanderthal range often leads to unequal introgression rates in Europe and Asia. Our result are thus in line with a recent paleogenetic study demonstrating that some hominin remains in the Altai Mountains are from Neanderthal type (18) and that Neanderthal range thus extended further east than previously believed. Although we have modeled the Asian range to extend up to the Altai region north of the Himalayas, we cannot be certain that the ancestors of East Asians migrated through this region. However, the facts that Papua New Guineans show signals of hybridization with another hominin (Denisovan) (2) and that their ancestors are likely to have followed a coastal southern route to the Pacific (19, 20) suggest that the Denisovan range must have extended more to the south and that the ancestors of East Asians may have indeed traveled north of the Himalayas, above the Denisovan range.
Although there are still clear uncertainties about the exact demographic history of Paleolithic humans and Neanderthals, it is important to realize that in our spatially explicit model of admixture, the final average levels of introgression increase logistically with the interbreeding success for various demographic scenarios (15) (Fig. S2). Even moderately larger interbreeding success rates would thus lead to extremely high levels of introgression. For instance, under scenario A, the expected level of Neanderthal introgression would already be ∼80% in contemporary Eurasians, with a hybrid interbreeding success of 5%. This therefore suggests that successful interbreeding must have been extremely limited between the two species.
It is also important to recognize that although we are calibrating our estimations by assuming that the inferred genome-wide average introgression rate is 1–3%, individual loci could show a large variance in introgression rate. For instance, under scenario A and for an interbreeding success rate of 0.5%, the distribution of introgression rates in the French and Chinese samples is very wide (Fig. S4), with some loci (7.4%) showing no introgression at all and others (7%) showing more than 20% Neanderthal introgression. This suggests that whole-genome data would need to be analyzed to infer introgression rates correctly but that introgression could be relatively easy to assess at some loci. Also of interest is the difference in introgression rate distributions between French and Chinese populations (Fig. S4), suggesting that the same locus could show drastically different introgression rates on the two continents.
Although we have not modeled this specific interaction, the larger extent of Denisovan introgression into Papua New Guineans (up to 5%) (2) could certainly fit a model of Papua New Guinean hybridization with Denisovans during their migration south of the Himalayas toward the Pacific. For comparison, an effective interbreeding success of 0.44% would indeed lead to an average 5% final Neanderthal introgression in Asians under scenario A. One can therefore imagine that similar figures would be obtained for Denisovan introgression if hybridization had occurred over a comparable range. Note that even lower interbreeding success rates would be compatible with 5% final introgression if hybridization had occurred over a longer period or a wider range.
Our model generally predicts an asymmetrical introgression from the local to the invading species for neutral markers (3, 15), which is precisely what was inferred from human-Neanderthal genetic comparisons (1). This asymmetry naturally occurs during a range expansion, but it is difficult to explain in the case of instantaneous hybridization without invoking some additional selective processes. Asymmetrical introgression proceeds because the invading population at the wave front becomes more and more admixed as it expands from the core region because of recurrent admixture events with the local population (15). Moreover, because most admixture events occur at or close to the wave front, where the invading local population is still growing, genes introgressing in the invading population are amplified by the population growth (3, 15). Therefore, a few introgression events can have a very large final impact. Under scenario A, as few as 197 introgression events lead, on average, to a 1% final introgression level, whereas 430 hybridization events are enough to reach a 3% final introgression level. These are very few successful interbreeding events, knowing that, under our model, about 1.875 million modern humans could have potentially interbred with Neanderthals. If we assume that both species interacted for 10,000 y during the range expansion of modern humans (11), successful admixtures between Neanderthals and humans would have occurred, on average, only once every 23–50 y over the whole hybrid zone to produce introgression levels of 1–3%, which shows that they were extremely rare events.
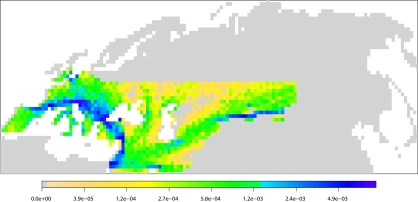
To understand the dynamics of the introgression process better, we show in Fig. 3 the spatial distribution of the introgression for scenario A assuming an interbreeding success rate of 0.5%. We see that introgression events can occur on almost the whole Neanderthal range but that there are clear geographical hot spots of introgression in the Middle East, on an expansion axis going from Anatolia to Spain, and in central Asia. Moreover, it should be obvious that introgression events have occurred mostly far away from the current location of the sampled populations and that Neanderthal lineages have been carried away by expanding populations accumulating introgressing lineages into regions where Neanderthals never existed (e.g., China, Papua New Guinea, North and South America). Because these introgression events always occurred on the human expansion wave front, it suggests that introgressed lineages found in Europeans and in Chinese should partly come from different Neanderthal populations present in introgression hot spots. If we divide the Neanderthal range into Europe, West Asia, and East Asia, we find that that 43%, 30%, and 27% of the introgression events can be assigned to these regions, respectively. Consequently, our model of introgression during range expansion would predict that Chinese and French populations should harbor both shared introgressed lineages (coming from Middle Eastern Neanderthals) and distinct introgressed lineages (coming from European and Asian Neanderthals). Our ability to recognize these different components of admixture from genomic data will depend on the extent of differentiation of Neanderthal populations living in these regions; however, even in the case of low levels of differentiation between Neanderthal populations (1, 2), different sections of the Neanderthal genome could still have introgressed into European and Asian populations or could show different rates of introgression (Fig. S4).
Fig. 3.
Spatial distribution of introgression events having left traces in the French and the Chinese populations obtained from 10,000 simulations of scenario A and assuming an interbreeding success rate of 0.5%. The color scale represents the estimated density of the introgression events per deme.
We would argue that our spatially explicit model of admixture provides a simple but powerful interpretative framework to explain interactions between modern humans and all other hominins. In particular, it is perfectly compatible with the observed Denisovan introgression in Papua New Guineans. If Denisovans had a large southwestern Asian range, Denisovan introgression would simply have occurred after previous episodes of Neanderthal introgression in the ancestors of Papua New Guineans expanding toward the Pacific along a southern Asian route. However, additional information about the spatial pattern of Denisovans would be necessary to model this specific interaction.
We show that a model of hybridization during range expansion can explain most patterns of human-Neanderthal ancestry (limited and relatively uniform introgression levels in Eurasia, introgression asymmetry, lack of mtDNA introgression, and signals of introgression in areas where Neanderthals never existed). Moreover, our model does not require postulation of an absence of admixture in Europe despite a documented and prolonged period of interaction, and it better fits the observed data than alternative spatially explicit scenarios of interbreeding occurring only in the Middle East. Finally, our model of continuous but limited interbreeding over the whole Neanderthal range predicts that the genomic components of Neanderthal introgression should be different in Europeans, northern Asians, and Papua New Guineans, because the ancestors of these populations would have admixed with potentially geographically and genetically distinct Neanderthal populations. It also implies that admixture events would have occurred hundreds of generations after the exit out of Africa. Improved analyses of Neanderthal sequences, of their genomic distribution in human populations, and of the estimation of admixture times should enable one to test this hypothesis.
Materials and Methods
Simulation Framework.
Using the program SPLATCHE2 (21), we simulated an expansion of early modern humans starting ∼50 kya (6) from a place arbitrarily located in northeastern Africa (Fig. 1, green dot) with an ancestral effective size of 50 individuals. Note that a different ancestral size or a different place of origin would not modify our results because neither affects the hybridization process occurring outside Africa, which is the relevant process in this study. At the onset of the modern human migration out of Africa, we assume that a large part of Europe and western Asia is already occupied by Neanderthals. We used two different potential maximum geographical ranges for the Neanderthals. In a first series of simulations, we considered that the Neanderthal range extended east up to the Altai Mountains in Central Asia, thus including the Okladnikov site located in the Altai region (18), and down to the shores of the Persian Gulf in the south (Fig. 1, brown, green, and violet areas). In a second series of simulations, we restricted the Neanderthal range only to the east to the Caucasus region, which corresponds to a more classic range based on archaeological remains (22) (Fig. 1, green and brown areas). In both cases, we restricted the range of Neanderthals to about 55° northern latitude, which corresponds approximately to the attested limit of Neanderthal northern extension (23). We also implemented scenarios in which hybridization between the two species was restricted to the Middle East (Fig. 1, brown area only). The continental surface of a Hammer–Aitoff projection of Eurasia and North Africa was divided in a grid of more than 6,000 geographical cells, each occupying an area of 100 × 100 km2, as shown in Fig. 1. Migrations are disallowed in cells located in seas as well as in those located in the Himalaya range. To test that deme area does not affect the results, we repeated the simulations with two different resolutions of deme areas (100 × 100 km2 and 50 × 50 km2), which seems appropriate for the simulation of the colonization of the old world by modern humans (24, 25), because it corresponds approximately to the territory of Mesolithic hunter-gatherers (26, 27). The density and migration parameters were scaled to the different resolution (a 4-fold difference for the densities and a 2.5-fold difference for the migration rates), such that population density per square kilometer remains the same in the two resolutions and the colonization times remain realistic (Table S1). Results obtained with the resolution of 50 × 50 km2 are presented in Figs. S1 and S3 and Table S1.
Each geographical cell belonging to the Neanderthal range contains two demes, one representing the Neanderthal population and the other one representing the H. sapiens population. All cells outside the Neanderthal range contain only one H. sapiens deme. The population density within each deme is logistically regulated using a model of density-dependent competition described elsewhere (3). This model is an extension of the Lotka–Volterra competition model, which includes intra- and interspecific competition; however, contrary to the classic Lotka–Volterra model, competition coefficients are not fixed but depend on the relative densities of both species in the cell, reflecting the fact that the influence of the members of one species on the other species grows with its density. If only one species is present, its density follows a simple logistic growth. Our model implies the progressive replacement of the species with the lower carrying capacity (i.e., Neanderthals) by the species with the higher carrying capacity (i.e., modern humans), as discussed below. We thus make the assumption that humans have a competitive edge over Neanderthals because of a better ability to exploit local resources (7).
Demographic Parameters Used in the Simulations.
For the 100 × 100 km2 deme area resolution, the carrying capacity of Neanderthals demes (KN) was set to a value corresponding to a density of about 0.025 individuals per 1 km2 (3), including juveniles and older nonreproducing people. Still in keeping with the study by Currat and Excoffier (3), and in agreement with density estimates for Pleistocene hunter-gatherers (28, 29), we used a fourfold higher density for modern humans (KH), which approximately corresponds to 0.1 individual per 1 km2. The K values used in the present study thus roughly correspond to those used in our previous simulation study (3), taking into account the fact that the number of gene copies of nuclear DNA is fourfold larger than for mtDNA. A growth rate (r) equal to 0.8 was set both for modern humans and Neanderthals, whereas migration rates were set to 0.1 for Neanderthals (mN) and to 0.2 for modern humans (mH). Those migration and growth parameters have been chosen because they result in the colonization of Europe in about 6,000 y, which fits well with archaeological data (11), and the arrival of modern humans in China between 45 and 40 kya, which is also coherent with archaeological information (7). Note that the colonization time of Europe in the current simulations is about twofold lower than that simulated in our previous study (3), so as to reflect the recent recalibration of 14C dates for the dispersal of modern humans over Europe (11). The choice of the shortest estimate for the colonization times is conservative regarding the estimation of the interbreeding success between Neanderthals and humans. Indeed, a lower interbreeding success rate is required to obtain an equivalent amount of Neanderthal ancestry if colonization time is longer. This combination of parameter values is referred to as the “standard” scenario A (Table 1). It results in an average of 11 generations of local cohabitations between Neanderthals and modern humans within each deme of a 100 × 100 km2 area, which fits well with the observation of a rapid local shift between Neanderthals and modern human that could have occurred locally over a few centuries (7). The parameters were scaled for the 50 × 50 km deme area resolution, as described above (Table S1).
Hybridization.
Hybridization between H. sapiens and Neanderthals may occur locally in all demes where individuals of both species coexist. Gene flow (hybridization) between the two species can only occur at geographical locations where they both coexist, and its amount is conditioned by the parameter γ, which represents an index of interbreeding success between the two species. The probability of a successful hybridization event, 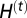 , at generation t is computed as the probability of locally drawing two individuals of different species multiplied by the interbreeding success rate of hybrids (γ) as
, at generation t is computed as the probability of locally drawing two individuals of different species multiplied by the interbreeding success rate of hybrids (γ) as
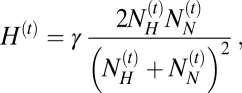 |
where 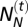 is the number of Neanderthals and
is the number of Neanderthals and 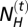 is the number of modern humans in the cell at generation t. It results in the introgression of
is the number of modern humans in the cell at generation t. It results in the introgression of 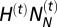 genes from Neanderthals to H. sapiens and
genes from Neanderthals to H. sapiens and 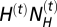 genes in the other direction at each generation t. For each tested scenario, we made 10,000 simulations for different values of γ, varying between 0 (no successful hybridization) and 1 (random mating between Neanderthals and modern humans). For each simulation, we recorded the amount of Neanderthal ancestry (i.e., Neanderthal genes introgressed into modern humans) in two contemporary human samples made up of 100 genes, one located in France and one located in China (Fig. 1, red dots). We then estimated the fraction of those simulations leading to a Neanderthal introgression level fitting the observations (i.e., between 1.9% and 3.1% of Neanderthal ancestry in contemporary samples). We slightly modified the program SPLATCHE2 (21) to output the number of hybridization events over the whole Neanderthal range and during the whole cohabitation period.
genes in the other direction at each generation t. For each tested scenario, we made 10,000 simulations for different values of γ, varying between 0 (no successful hybridization) and 1 (random mating between Neanderthals and modern humans). For each simulation, we recorded the amount of Neanderthal ancestry (i.e., Neanderthal genes introgressed into modern humans) in two contemporary human samples made up of 100 genes, one located in France and one located in China (Fig. 1, red dots). We then estimated the fraction of those simulations leading to a Neanderthal introgression level fitting the observations (i.e., between 1.9% and 3.1% of Neanderthal ancestry in contemporary samples). We slightly modified the program SPLATCHE2 (21) to output the number of hybridization events over the whole Neanderthal range and during the whole cohabitation period.
Alternative Scenarios.
To evaluate the effects of different demographic parameter on our results, we explored 10 alternative scenarios defined by setting each time parameter to an extreme value, although still preserving realistic colonization times. We used either a larger (Table S1, scenario B) or smaller (Table S1, scenarios C and C′) carrying capacity than for the basic scenario A, leading to a range of density between 0.0125 and 0.2 individuals per 1 km2 for H. sapiens and between 0.003 and 0.05 individuals per 1 km2 for Neanderthals We also performed simulations using either a migration rate 50% smaller than in scenario A (scenario D) or a growth rate also reduced to 50% of its value in scenario A (scenario E). We considered possible variability in H. sapiens and Neanderthal deme densities to reflect potential habitat heterogeneity. We did this by using five different K values only for modern humans (200, 400, 800, 1,200, and 1,600), distributed regularly over the whole area with the condition that neighboring demes never have an identical carrying capacity (scenario F). For this latter scenario, the average carrying capacity is close to that modeled under scenario A (840 vs. 800). Then, based on scenario F, we made the K values of Neanderthals also covary with the K values of modern humans (e.g., so that the ratio KH/KN remains constant). This scenario F′ assumes that landscape heterogeneity of resources exists but that humans were systematically better at exploiting them than Neanderthals, such that KH and KN are fully correlated over space. Contrastingly, scenario F′′ assumes that both species exploit different local resources and that there is no correlation in the densities of species over space. In that case, we used 80 different combinations of random KH (between 200 and 1,600) and KN (between 50 and 400) values distributed arbitrarily over the map. Finally, we also simulated two scenarios where both human and Neanderthal densities are higher in the Middle East (Fig. 1, brown area) than in the rest of their range. For scenario G, KN = 200 and KH = 800 in the Middle East and KN = 50 and KH = 200 in the rest of the map. For scenario G′, the carrying capacities in the Middle East are identical to those in scenario G but KN = 100 and KH = 400 everywhere else.
The goodness of fit of alternative scenarios to the data was compared by means of the AIC (16), defined as 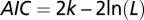 , where k is the number of estimated parameters (here, k = 1) and L is the maximum likelihood of the model. The probabilities of each model, m, relative to the standard model A were estimated as the weighted AIC,
, where k is the number of estimated parameters (here, k = 1) and L is the maximum likelihood of the model. The probabilities of each model, m, relative to the standard model A were estimated as the weighted AIC, 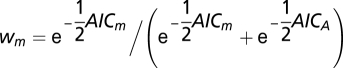 (30), and the relative probability of model A was compared with that of model m as
(30), and the relative probability of model A was compared with that of model m as 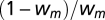 , sometimes called the evidence ratio in favor of model A. Models that differ in the AIC by less than 2 are generally considered to be equally well supported by the data, which corresponds to an evidence ratio of 2.7.
, sometimes called the evidence ratio in favor of model A. Models that differ in the AIC by less than 2 are generally considered to be equally well supported by the data, which corresponds to an evidence ratio of 2.7.
Supplementary Material
Acknowledgments
We are grateful to Mark Beaumont, Montgomery Slatkin, and François Balloux for their helpful comments on the manuscript. The comments of two anonymous reviewers were also very helpful in clarifying several important points. This study was supported by Swiss National Science Foundation Grants 31003A-127465 (to M.C.) and 3100A0-126074 (to L.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107450108/-/DCSupplemental.
References
- 1.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reich D, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Currat M, Excoffier L. Modern humans did not admix with Neanderthals during their range expansion into Europe. PLoS Biol. 2004;2:e421. doi: 10.1371/journal.pbio.0020421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serre D, et al. No evidence of Neandertal mtDNA contribution to early modern humans. PLoS Biol. 2004;2:E57. doi: 10.1371/journal.pbio.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krings M, et al. Neandertal DNA sequences and the origin of modern humans. Cell. 1997;90:19–30. doi: 10.1016/s0092-8674(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 6.Klein RG. Darwin and the recent African origin of modern humans. Proc Natl Acad Sci USA. 2009;106:16007–16009. doi: 10.1073/pnas.0908719106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein RG. Out of Africa and the evolution of human behavior. Evol Anthropol. 2008;17:267–281. [Google Scholar]
- 8.Eswaran V, Harpending H, Rogers AR. Genomics refutes an exclusively African origin of humans. J Hum Evol. 2005;49:1–18. doi: 10.1016/j.jhevol.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Hawks J, Cochran G, Harpending HC, Lahn BT. A genetic legacy from archaic Homo. Trends Genet. 2008;24:19–23. doi: 10.1016/j.tig.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Hoffecker JF. Out of Africa: Modern human origins special feature: The spread of modern humans in Europe. Proc Natl Acad Sci USA. 2009;106:16040–16045. doi: 10.1073/pnas.0903446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellars P. A new radiocarbon revolution and the dispersal of modern humans in Eurasia. Nature. 2006;439:931–935. doi: 10.1038/nature04521. [DOI] [PubMed] [Google Scholar]
- 12.Mellars P. Why did modern human populations disperse from Africa ca. 60,000 years ago? A new model. Proc Natl Acad Sci USA. 2006;103:9381–9386. doi: 10.1073/pnas.0510792103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mellars P. Neanderthals and the modern human colonization of Europe. Nature. 2004;432:461–465. doi: 10.1038/nature03103. [DOI] [PubMed] [Google Scholar]
- 14.Nordborg M. On the probability of Neanderthal ancestry. Am J Hum Genet. 1998;63:1237–1240. doi: 10.1086/302052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Currat M, Ruedi M, Petit RJ, Excoffier L. The hidden side of invasions: Massive introgression by local genes. Evolution. 2008;62:1908–1920. doi: 10.1111/j.1558-5646.2008.00413.x. [DOI] [PubMed] [Google Scholar]
- 16.Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–723. [Google Scholar]
- 17.Fabre V, Condemi S, Degioanni A. Genetic evidence of geographical groups among Neanderthals. PLoS ONE. 2009;4:e5151. doi: 10.1371/journal.pone.0005151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krause J, et al. Neanderthals in central Asia and Siberia. Nature. 2007;449:902–904. doi: 10.1038/nature06193. [DOI] [PubMed] [Google Scholar]
- 19.Macaulay V, et al. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science. 2005;308:1034–1036. doi: 10.1126/science.1109792. [DOI] [PubMed] [Google Scholar]
- 20.Bulbeck D. Where river meets sea—A parsimonious model for Homo sapiens colonization of the Indian Ocean rim and Sahul. Curr Anthropol. 2007;48:315–321. [Google Scholar]
- 21.Ray N, Currat M, Foll M, Excoffier L. SPLATCHE2: A spatially-explicit simulation framework for complex demography, genetic admixture and recombination. Bioinformatics. 2010;26:2993–2994. doi: 10.1093/bioinformatics/btq579. [DOI] [PubMed] [Google Scholar]
- 22.Klein RG. Paleoanthropology. Whither the Neanderthals? Science. 2003;299:1525–1527. doi: 10.1126/science.1082025. [DOI] [PubMed] [Google Scholar]
- 23.Hublin JJ. Out of Africa: Modern human origins special feature: The origin of Neandertals. Proc Natl Acad Sci USA. 2009;106:16022–16027. doi: 10.1073/pnas.0904119106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Currat M, Excoffier L. The effect of the Neolithic expansion on European molecular diversity. Proc Biol Sci. 2005;272:679–688. doi: 10.1098/rspb.2004.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ray N, Currat M, Berthier P, Excoffier L. Recovering the geographic origin of early modern humans by realistic and spatially explicit simulations. Genome Res. 2005;15:1161–1167. doi: 10.1101/gr.3708505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ammerman A, Cavalli-Sforza LL. The Neolithic Transition and the Genetics of Populations in Europe. Princeton: Princeton Univ Press; 1984. p. 176. [Google Scholar]
- 27.Gronenborg D. A variation on a basic theme: The transition to farming in southern central Europe. J World Prehist. 1999;13:123–210. [Google Scholar]
- 28.Bocquet-Appel J-P, Demars PY. Population kinetics in the Upper Paleolithic in Western Europe. J Archaeol Sci. 2000;27:551–570. [Google Scholar]
- 29.Steele J, Adams JM, Sluckin T. Modeling Paleoindian dispersals. World Archaeol. 1998;30:286–305. [Google Scholar]
- 30.Burnham KP, Anderson DR. Mathematical models for nonparametric inferences from line transect data. Biometrics. 1976;32:325–336. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.