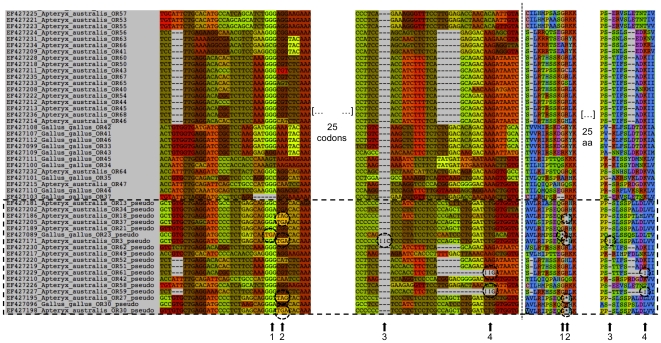
Figure 2. Snapshots of a multiple alignment of 93 functional and 18 pseudogene sequences from brown kiwi (Apteryx australis) and domestic chicken (Gallus gallus) olfactory repertoires.
The same alignment region is displayed at the NT (left) and AA (right) levels. The 18 pseudogene sequences are boxed. Stop codons (stars in amino acid sequences) occurring at sites 1 and 2, and frameshifts (exclamation marks) inferred by MACSE at sites 3 and 4 are circled. MACSE guideline parameters for pseudogene datasets were used (see Fig. 1 for details.)