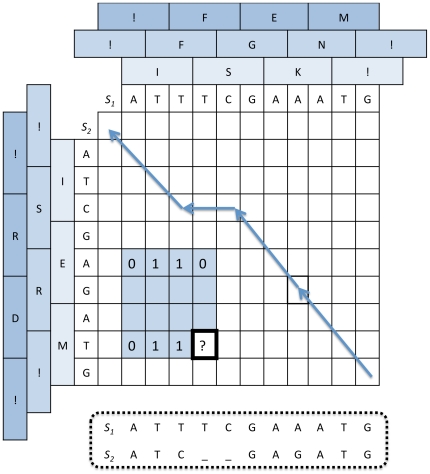
Figure 6. Alignment of two DNA coding sequences.
Like for classical Needleman-Wunsch, an array is used to store the cost of an optimal alignment between prefixes of  ( = ATTTCGAAATG) and prefixes of
( = ATTTCGAAATG) and prefixes of 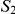 ( = ATCGAGATG). The AA translations of those sequences are used to detect STOP codons and to evaluate codon substitutions based on their AA translations. The value of each cell is computed using 15 nearby cells. For instance, the bold cell value is computed based on its 15 colored neighbors. Among those 15 cells, some induced frameshifts in one or both sequences (see Fig. 7 for details). For instance cells marked with a “0” cause no frameshift, those marked by “1” cause a frameshift for
( = ATCGAGATG). The AA translations of those sequences are used to detect STOP codons and to evaluate codon substitutions based on their AA translations. The value of each cell is computed using 15 nearby cells. For instance, the bold cell value is computed based on its 15 colored neighbors. Among those 15 cells, some induced frameshifts in one or both sequences (see Fig. 7 for details). For instance cells marked with a “0” cause no frameshift, those marked by “1” cause a frameshift for 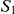 but not for
but not for 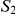 . The optimal path (indicated by arrows) is determined using a backtracking process similar to the classical one, except that 15 possible moves are now considered. The alignment corresponding to this arrow path is depicted in the dashed box.
. The optimal path (indicated by arrows) is determined using a backtracking process similar to the classical one, except that 15 possible moves are now considered. The alignment corresponding to this arrow path is depicted in the dashed box.