Figure 1.
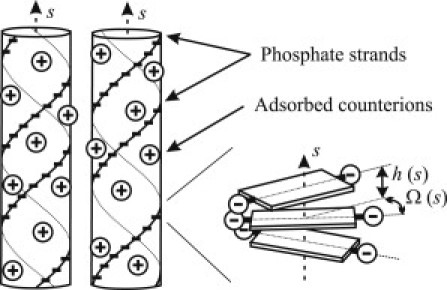
Electrostatic sequence homology recognition and pairing of parallel DNA double helices. Alignment of negatively charged phosphate strands opposite to positively charged counterions in the grooves of the apposing molecule reduces the electrostatic energy. This may lead to intermolecular attraction and pairing, provided that a sufficient fraction of phosphate charges is compensated by bound counterions. Sequence-related variations in the twist Ω(s) and axial rise h(s) per basepair (depicted on the right) lead to variations in the helical pitch. These disrupt the energetically favorable alignment of two apposing molecules with nonhomologous sequences.
