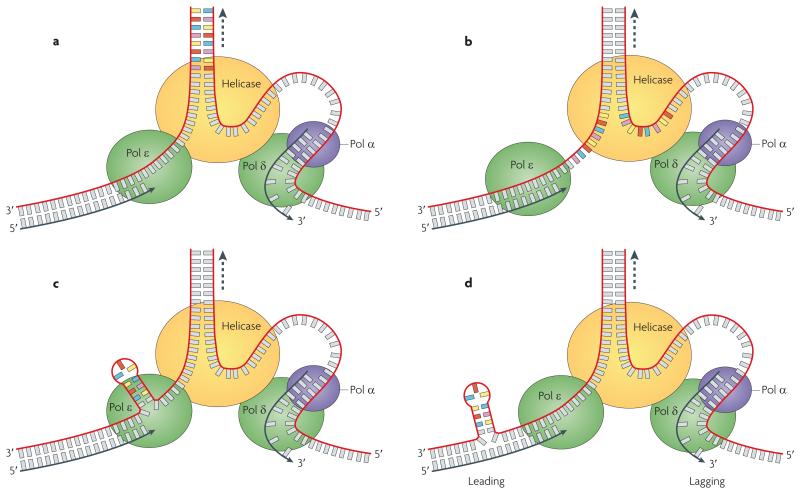
Figure 4. Slippage model for small trinucleotide repeat length changes.
Illustrated is a slippage error on the leading strand template. a | Before reaching the CAG repeat sequence (coloured bars), the replicative DNA polymerase (Pol ε on the leading strand) and DNA helicase are tightly coupled and coordinate with each other at the replication fork. b | When the polymerase encounters the CAG tract, polymerase progression is slowed but the helicase speed is unaffected (space between green and yellow ovals). c | To avoid uncoupling of the polymerase and the helicase, the leading strand polymerase bypasses a segment of unreplicated CAG template on the leading strand. d | Coupling stress is relieved when the fork has moved through the CAG repeat region and resumes copying random DNA sequence. Base pairing and processing of the looped strand results in instability of the CAG tract (not shown). Deletions occur if the loops form on the template strand and insertions occur when loop formation is on the daughter strand (not shown). Slippage can occur in both leading and lagging strand orientation, although instability (most often involving deletions) is highest when the repeats are on the lagging strand template (Pol α is the primase that synthesizes primers on the lagging strand). Figure is modified, with permission, from ref. 86 © (2008) The American Society for Biochemistry and Molecular Biology.