Abstract
We have previously shown that the transcription-promoting activity of serum response factor (SRF) is partially regulated by its extranuclear redistribution. In this study, we examined the cellular mechanisms that facilitate SRF nuclear entry in canine tracheal smooth muscle cells. We used in vitro pull-down assays to determine which karyopherin proteins bound SRF and found that SRF binds KPNA1 and KPNB1 through its nuclear localization sequence. Immunoprecipitation studies also demonstrated direct SRF–KPNA1 interaction in HEK293 cells. Import assays demonstrated that KPNA1 and KPNB1 together were sufficient to mediate rapid nuclear import of SRF-GFP. Our studies also suggest that SRF is able to gain nuclear entry through an auxiliary, nuclear localization sequence–independent mechanism.
Keywords: serum response factor, nuclear import, karyopherin
Clinical Relevance
Serum response factor is a potent transcription factor that activates the promoters of several smooth muscle–specific and immediate early genes. Our findings suggest that the formation of the ternary complex, KPNA1-KPNB1-SRF, is the primary mechanism by which serum response factor (SRF) gains nuclear entry. Additional studies of SRF nuclear export may help determine whether its nuclear-cytoplasmic shuttling is altered in disease states such as asthma, atherosclerotic heart disease, or pulmonary fibrosis, where SRF might play an important role in pathogenesis.
Serum response factor (SRF) binds and activates the promoter of several smooth muscle specific genes, such as those encoding SM22α and smooth muscle myosin heavy chain, as well as the promoter of several immediate early genes, including c-fos. We have previously shown that the transcription-promoting activity of SRF is partially regulated in cultured smooth muscle cells by its extranuclear redistribution (1). The pathways that regulate the subcellular distribution of SRF have not been fully elucidated but appear to include Rho kinase– and Akt-dependent mechanisms. Inhibition of Rho kinase with Y-27632 decreased SRF-dependent transcription, in part by cytoplasmic redistribution of SRF out of the nucleus of cultured airway myocytes (2). PDGF-BB treatment stimulated the extranuclear redistribution of SRF in vascular myocytes through PI-3 kinase α– and Akt-dependent signaling (3). Movement of SRF out the nucleus has also been observed when NIH3T3T cells terminally differentiate into adipocytes (4). Furthermore, hemaglutinin-tagged SRF disappeared from fibroblast nuclei within 15 minutes after nuclear import was inhibited (5). Despite these clear demonstrations that the subcellular localization of SRF is dynamic and physiologically regulated and that this process partially controls SRF-dependent gene expression, very little is known about the detailed cellular mechanisms that facilitate SRF nuclear entry.
Over the last 20 years, many details of the nuclear import process have been elucidated. Small molecules (< 40 kD) are able to freely pass from the cytoplasm into the nucleus (and vice versa) through the nuclear pore complex (NPC). However, the nuclear trafficking of molecules larger than 40 kD is physiologically regulated. The NPC is comprised of multiple proteins (6)—as many as 30 different species according to some estimates—that form a channel through the nuclear envelope and allow for bidirectional movement of proteins and RNAs. Most proteins that require nuclear entry, such as transcription factors, contain a nuclear localization sequence (NLS). The classical NLS is a short stretch of 5 to 10 amino acids containing several basic residues (arginine and/or lysine), whereas the bipartite NLS is comprised of two short stretches of basic amino acids interrupted by a 10- to 12-amino acid linker. Importin α and β proteins have been identified as the major nuclear transporters that are capable of binding the NLS. In most cases, α importins act as adaptor proteins, linking cytoplasmic cargoes to importin β, which in turn binds the NPC and effects nuclear entry. The N-terminus of α importins functions as the importin β-binding domain (IBB), and the central NLS-binding region of α importins is composed of a series of armadillo repeats that are able to bind two classical NLS proteins or one bipartite NLS protein (7). There is some evidence that the N-terminal IBB can bind the NLS of α importins when it is not bound to an import substrate, thus acting in an autoinhibitory fashion (8). When an importin α is bound to importin β and an NLS-containing import substrate, the aggregate is often referred to as the ternary complex. Upon entering the nucleus, the ternary complex is dissociated after Ran-GTP, a small nuclear GTPase, binds to importin β. Thus, the Ran-GTP–rich environment of the nucleus favors ternary complex dissociation, whereas ternary complex formation occurs in the Ran-GDP–rich environment of the cytoplasm.
SRF, with apparent molecular weight of 67 kD, requires coordinated nuclear import through the NPC to gain nuclear access and bind (as a homodimer) to its consensus DNA sequence CC[AT]6GG, or “CArG box” (9). Although a functional classical NLS was identified at amino acids 95 to 100 (5), other details concerning SRF nuclear transport remain unknown. In this study, we provide evidence that SRF can enter the nucleus through a mechanism that requires importin α1 (KPNA1) and importin β (KPNB1). Because there is confusion in the literature concerning the naming of various importin α species, hereafter we refer to all importin proteins by their karyopherin (KPN) designations.
Materials and Methods
Plasmid construction, buffer composition, SRF functional domains, and SRF constructs are described in the online supplement.
In Vitro Binding Assay
Glutathione-S-transferase (GST)-fusion proteins expressed in Escherichia coli were bound to Glutathione Sepharose 4B beads (Amersham Biosciences, Piscataway, NJ). 35S-methionine–labeled karyopherin or SRF proteins were synthesized using a coupled in vitro transcription/translation kit (Promega, Madison, WI). 35S-labeled proteins were incubated with beads coated with GST, GST-wtSRF, GST-mutant SRF, or GST-karyopherin for 4 hours at 4°C with continuous rocking in protein interaction buffer. Beads were pelleted and washed five times in protein interaction buffer, and then bound 35S-labeled proteins were eluted by boiling for 5 minutes in Laemlli's sample buffer, size fractionated by SDS-PAGE, and autoradiographed.
Subcellular Localization of EGFP-SRF Fusion Proteins
Canine tracheal smooth muscle (CTSM) cells of passage 1 or 2 were grown to 70% confluence on glass cover slips in 12-well plates (1). pEGFP-SRF plasmids were transiently transfected in Optimem (Life Technologies, Carlsbad, CA) using 1.2 μg DNA and 6 μg LipofectAMINE (Life Technologies) per well (final concentration of 0.012 μg/μl per manufacturer's recommendations) (2). After 5 hours, cells were re-fed medium containing 10%FCS for 24 hours. Coverslips were washed consecutively in cytoskeleton buffer (CB) containing 3% paraformaldehyde and 0.1% Triton X-100 for 5 minutes, in CB twice for 5 minutes, in CB with 3% paraformaldehyde for 15 minutes, and in CB twice for 5 minutes, then stored in CB at 4°C. Nuclei were stained with Hoechst 33342. Cellular distribution of EGFP-SRF fusion proteins and Hoechst 33342 were assessed by fluorescence microscopy at 40×. Some CTSM were treated with 10 nM of leptomycin for 24 hours before imaging.
Immunoprecipitation
Immunoprecipitation methods were described previously (10).
Import Assay
CTSM cells were grown on glass cover slips to approximately 80% confluence. Cells were washed with transport buffer (TB) twice before incubation with digitonin (50 μg/μl) for 1.5 minutes on ice. Permeabilized cells were washed with TB three times and then incubated in a room–temperature, high-humidity chamber with 100 μl import buffer (10 μl ATP regenerating system, 3 μg SRF-GFP, 2 μM karyopherin protein, and TB to final volume). Real-time images of unfixed cells were taken every 60 seconds for 6 minutes.
Results
SRF Can Enter the Nucleus Despite Mutation of its Dimerization Domain, DNA-Binding Domain, or NLS
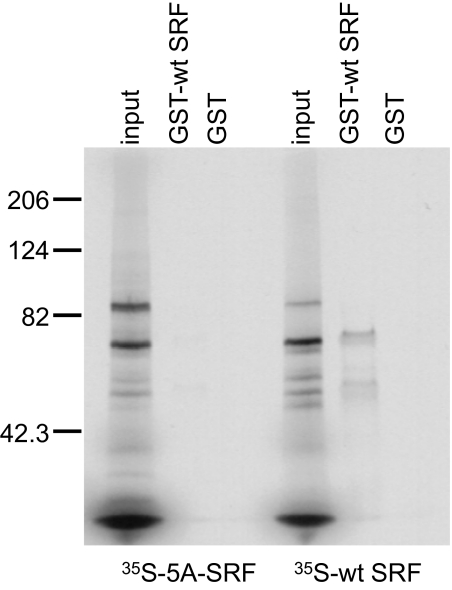
Although SRF dimerization is required for binding to its cognate site on DNA (11), it is not known whether SRF dimerization is also required for nuclear entry. The crystal structure of SRF bound to DNA also predicts that the hydrophobic span in the βI sheet of the dimerization domain (183-VLLLV-187) is involved in homodimer formation of SRF (12). We posited that disruption of the βI sheet should therefore also prevent dimer formation. To test this possibility, we constructed the 5A-SRF mutant, in which 183-AAAAA-187 replaces the wild-type (wt) sequence, and used a GST pull-down assay to assess its potential for dimerization with wt SRF. As shown in Figure 1, GST-wt SRF specifically binds to 35S-labeled wt SRF, but binding to the 35S-labeled 5A-SRF mutant is greatly reduced, as reflected in the presence of a very faint band evident in the 5A-SRF lane.
Figure 1.
Binding of the dimerization mutant 35S-5A serum response factor (SRF) to wild-type (wt)-SRF in solution is dramatically reduced but not eliminated. In vitro pull-down assay reveals that glutathione-S-transferase (GST)-wt SRF binds 35S-wt SRF much more avidly than it binds 35S-5A-SRF. Images are representative of at least two separate experiments and display the 35S-SRF species pulled down with GST-wt SRF. The smaller band in the GST-wt SRF lane represent nonspecific binding of 35S-wt SRF to the GST-wt SRF beads or breakdown products of 35S-wt SRF. GST-containing beads were used as the negative control for each pull-down assay. Five microliters of TNT transcription/translation solution, containing 35S-wt-SRF or 35S-5A-SRF proteins were used for each binding assay. Equal volumes (3 μl) of input proteins are shown for each binding assay.
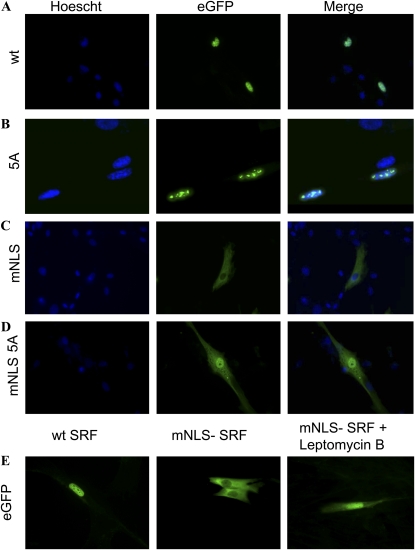
To determine whether SRF can enter the nucleus despite NLS, dimerization, and DNA binding mutations, we transfected CTSM cells with plasmids encoding wt or mutant EGFP-SRF fusion proteins and assessed their subcellular distributions with fluorescence microscopy. EGFP-wt SRF gains nuclear entry and is found in a distribution corresponding to that expected for chromatin (Figure 2A). The dimerization mutant EGFP-5A-SRF also gains nuclear entry, but within the nucleus accumulates in discrete collections that do not reflect any obvious nuclear structure (Figure 2B). We speculated that the abnormal distribution within the nucleus of dimerization SRF reflects its inability to bind to DNA (11) and therefore assessed the distribution of EGFP-pm1-SRF, in which the SRF moiety contains point mutations outside the dimerization domain that prevent DNA binding (13) but do not inhibit dimerization. Like the dimerization mutant EGFP-5A-SRF, EGFP-pm1-SRF accumulates in discrete intranuclear collections (data not shown). Together, these findings demonstrate that SRF can enter the nucleus despite mutations that prevent its DNA binding (pm1) or greatly reduce its dimerization (5A), presumably also reducing DNA binding, and suggest that lack of DNA binding leads to its accumulation in discrete intranuclear collections.
Figure 2.
SRF is able to enter the nucleus despite mutation of its dimerization domain or NLS. Intracellular distributions of (A) wt SRF, (B) dimerization 5A SRF mutant, (C) mNLS SRF, and (D) mNLS/5A double mutant SRF are shown. Images displayed are nuclear (Hoechst) stained, GFP alone (revealing SRF localization), and both images merged. (E) Intracellular distributions of wt-SRF and mNLS-SRF in the absence of treatment and of mNLS-SRF during treatment with leptomycin B. For all images, canine tracheal smooth muscle cells were transfected with EGFP-wt– or EGFP-mutant–SRF plasmids before fixation and imaging with fluorescence microscopy. Experiments were repeated four times, with representative images shown at 400×. Images in B are at higher magnification (∼ 1,000×) to better demonstrate the intranuclear distibution of 5A-SRF.
As expected (5), mutation of amino acids 95 and 96 in the SRF nuclear localization sequence (RR → EE to generate EGFP-mNLS-SRF) prevented nuclear accumulation of EGFP-mNLS-SRF (Figure 2C) under normal circumstances. This implies that either: 1) EGFP-mNLS-SRF is completely unable to enter the nucleus because its NLS mutation obliterates the only operative mechanism for its nuclear import; or 2) the nuclear export of EGFP-mNLS-SRF is more efficient than any nuclear import that might occur despite mutation of its NLS, thereby shifting the balance between nuclear import and nuclear export as to prevent significant nuclear SRF accumulation. To distinguish between these two possibilities, we treated EGFP-mNLS-SRF–transfected cells with 10 nM leptomycin B, which inhibits nuclear export. Figure 2E demonstrates nuclear accumulation of some EGFP-mNLS-SRF when nuclear export is inhibited. This result indicates that SRF possesses an auxiliary nuclear import mechanism that does not rely upon an intact NLS at aa 95 to 100. We entertained the possibility that, as one potential auxiliary nuclear import mechanism, EGFP-mNLS-SRF gains nuclear entry by dimerizing with endogenous wt SRF, riding “piggyback” on wt SRF as it traverses the nuclear pore. To test this possibility, we constructed a doubly mutant EGFP-SRF, in which the SRF moiety contains the mNLS and dimerization blunting 5A mutations. When expressed in CTSM cells, EGFP-mNLS-5A-SRF accumulated in the nucleus of some cells (Figure 2D). Thus, neither an intact NLS nor an intact dimerization domain is required for SRF nuclear entry.
KPNA1 and KPNB1 Bind SRF In Vitro, and Mutation of the NLS Abolishes This Interaction
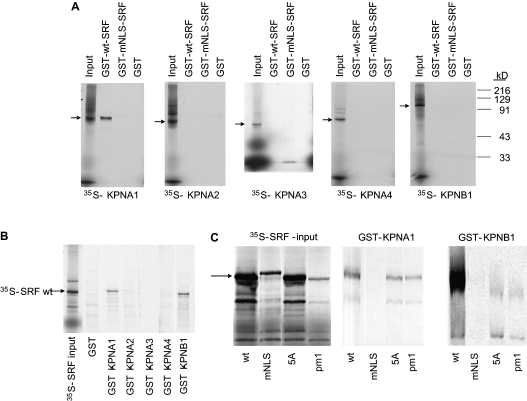
To determine which karyopherins mediate SRF nuclear entry, we first evaluated their abilities to bind SRF in vitro by GST pulldown assay. In these studies, GST-SRF specifically bound 35S-KPNA1 but not radiolabeled KPNA2, -3, or -4, or 35S-KPNB1 (Figure 3A). The addition of unlabeled KPNB1 to the 35S-KPNA2–4 reactions did not result in additional binding of GST-SRF (data not shown), indicating that the lack of SRF binding to these α karyopherins did not reflect a requirement for ternary complex formation. Moreover, mutation of the SRF NLS abrogates binding, as would be expected for an interaction between a classical NLS and a karyopherin α adaptor (Figure 3A). We further examined SRF–KPNA interactions by generating GST-KPN–coated beads and testing their abilities to pull down 35S-SRF. Once again, SRF interacted with KPNA1, for GST-KPNA1 beads pulled down 35S-SRF, but not 35S-mNLS-SRF (Figures 3B and 3C). SRF mutations that prevent DNA binding (pm1) or dimerization (5A) did not inhibit this interaction (Figure 3C), further supporting the notion that SRF binds to KPNA1 through its NLS.
Figure 3.
KPNA1 and KPNB1 bind SRF in vitro and NLS mutation abolishes this interaction. (A) In vitro pull-down assay reveals that GST-SRF binds 35S-KPNA1, and mutation of the NLS abolishes this interaction. GST-SRF does not bind 35S-KPNA2–4 or 35S-KPNB1. (B) Pull-down assay reveals that GST-KPNA1 and GST-KPNB1 bind SRF, whereas GST-KPNA2–4 have no significant binding. (C) Pull-down assay demonstrates that a mutation that disrupts the SRF NLS (mNLS), but not mutations that disrupt SRF DNA-binding (pm1) or dimerization (5A), prevents binding to KPNA1 and to KPNB1. Images are representative of at least two separate experiments.
GST-KPNB1 beads specifically bound to and pulled down wt 35S-SRF (Figure 3B). As with the SRF–KPNA1 interaction, mutation of the SRF NLS prevented the SRF–KPNB1 interaction (Figure 3C). Although there is variability in the input strength for the 35S-SRF proteins, there is clear binding of GST-KPNA1 and GST-KPNB1 by SRF mutant pm1 (which had the weakest input) and by SRF mutant 5A. Therefore, neither the DNA binding (pm1) nor dimerization (5A) mutants inhibited the interaction of SRF and KPNA1 or KPNB1. Thus, it appears that SRF can also directly bind to KPNB1 through its NLS. Direct interaction between KPNB1 and other nuclear cargoes (e.g., Smad3 [14]) has been reported previously.
KPNA1 Mediates Nuclear Import of SRF In Vivo
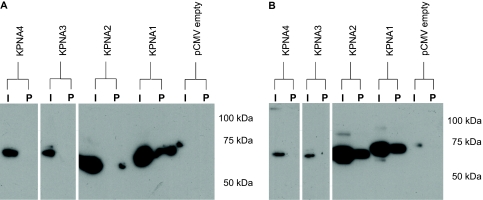
To determine the principal karyopherin species responsible for SRF nuclear entry in vivo, we first explored interactions between KPNA1–4 and SRF in living cells. HEK293 cells were transiently transfected with pSRF-HA and pKPNA1–4-myc plasmids, and then immunoprecipitation was performed using nuclear and cytoplasmic extracts to assess in vivo interaction. Pull-down with anti-HA antibody–coated beads (pulls down SRF) followed by Western blotting with anti-myc antibody (detects karyopherin) revealed that SRF interacts in vivo with KPNA1 in the nucleus but not with KPNA2–4 in this compartment (Figure 4A). KPNA2 does interact with SRF in the cytosol (Figure 4B). Given that SRF binds to KPNA1 and that KPNA proteins are known to bind KPNB1, we did not probe membranes for KPNB1 because demonstration of KPNB1 in the immunoprecipitate would not provide evidence for or against direct SRF–KPNB1 interaction in vivo.
Figure 4.
HA-SRF binds KPNA1-myc and KPNA2-myc in vivo. (A) Immunoprecipitation of nuclear extract with anti-HA beads (pulls down SRF) and Western blot with anti-myc antibody (karyopherin) demonstrates in vivo interaction of SRF with KPNA1 but not with KPNA2–4 in transfected 293 cells. Equal amounts of nuclear protein were loaded in each input (I) lane. (B) Immunoprecipitation of cytoplasmic proteins with anti-HA beads (pulls down SRF) and Western blot with anti-myc antibody (karyopherin) demonstrates in vivo interaction of SRF with KPNA1 and KPNA2 but not with KPNA3–4 in cytosolic compartment of transfected 293 cells. Equal amounts of total protein were loaded in each input (I) lane. P refers to the pellet that is pulled down by the antibody-coated beads.
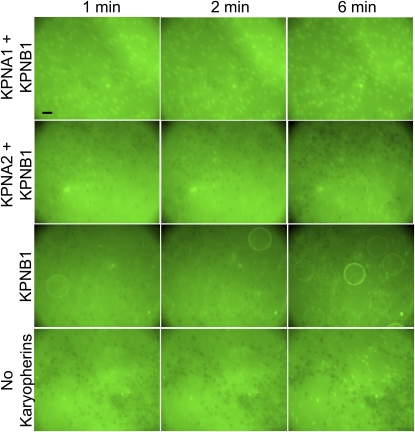
Next, an import assay was performed to determine which karyopherin proteins can mediate nuclear import of SRF-EGFP in digitonin-permeabilized CTSM cells. Pictures were taken every minute; representative images are presented in Figure 5. The addition of KPNA1 and KPNB1 together to the import buffer resulted in rapid entry of SRF-EGFP into the nucleus of nearly every permeabilized cell. In contrast, KPNA2 and KPNB1 together did not facilitate much nuclear SRF-EGFP accumulation. Most of the nuclei are visualized as “punched out” areas, devoid of the surrounding green SRF-EGFP protein that is unable to gain nuclear access. KPNB1 alone, in the absence of any KPNA protein, did not mediate substantial nuclear entry of SRF-EGFP, nor did nuclear accumulation of SRF-EGFP occur in most cells when all karyopherin species were excluded from the import buffer (the negative control condition). A few permeabilized cells accumulated SRF-EGFP after 6 minutes of import; this slow nuclear accumulation of SRF-EGFP might have resulted from damage to the nuclear envelope during digitonin permeabilization, but it also raises the possibility that a secondary karyopherin-independent import mechanism exists. Together, these data indicate that KPNA1 and KPNB1 primarily act in concert to effect SRF nuclear entry.
Figure 5.
KPNA1 and KPNB1 mediate the nuclear import of SRF-GFP in digitonin-permeabilized CTSM. The bright green background in these images is fluorescent protein (SRF-GFP) added to the artificial cytoplasm. The “punched out” areas with much less green fluorescence represent nuclei into which nuclear import of SRF-GFP has not occurred. Neither KPNA2 and KPNB1, KPNB1 alone nor no kayopherins effect much SRF-GPF nuclear import. The large circles with fluorescent edges in the KPNB1 images are air bubbles. The reference bar is 20 μm in length. Images are representative of at least three separate experiments.
Discussion
New details of SRF nuclear import are reported in this study. First, SRF is able to enter the nucleus despite mutation that markedly reduces dimerization. Previous studies demonstrated that SRF must bind its DNA consensus sequence as a homodimer to realize its transcription-promoting activity and that SRF exists as a dimer in solution (11). Nevertheless, an SRF mutation that significantly reduces dimerization (EGFP-5A-SRF) enters the nucleus (see Figure 2B). In contrast, mutations that inhibit dimer formation of STAT1 and STAT2—transcription factors that also bind DNA as dimers—prevent their nuclear import and result in the cytoplasmic accumulation of STAT monomers (15). Thus, although strong dimerization is a requirement for the nuclear import of some transcription factors that act as dimers, this requirement does not appear to hold for SRF.
Our study shows that SRF binds KPNA1 and KPNB1 through its NLS. Gauthier-Rouviere and colleagues demonstrated that SRF contains a classical NLS at amino acids 95 to 100 (RRGLKR) and that mutation of amino acids 95 and 96 (RR → EE) prevents the nuclear accumulation of SRF (5). We have demonstrated that SRF binds two karyopherin proteins, KPNA1 and KNPB1, through its NLS in vitro. Acting as adaptors, KPNA proteins facilitate nuclear entry by binding KPNB1, which in turn binds the NPC and effects nuclear entry of the ternary complex. Nevertheless, several proteins, including Smad-3, CREB, GAL4, cyclin-B1, and parathyroid hormone-related protein, bind directly to KPNB1 and thus do not require KPNA adaptors to gain nuclear entry (14, 16–19). Our binding assays demonstrate that GST-SRF protein bound to glutathione beads does not bind 35S-KPNB1, whereas 35S-SRF does bind glutathione-bound GST-KPNB1. These disparate results are likely a result of steric hindrance. KPNB1 is comprised of 19 tandem HEAT repeats, each one comprised of two connected helices. The IBB domain, present in KPNA proteins, has been shown to bind HEAT repeats 7 to 19 in the C-terminal portion of KPNB1, whereas parathyroid hormone-related protein and SREBP-2 directly bind the inner concave surface of the KPNB1 superhelix (20). We speculate that the glutathione bead–GST–SRF complex is too big to gain access to the inner concave surface of KPNB1 and thus cannot bind directly to 35S-KPNB1. When 35S-SRF is used in the binding assay, its NLS is able to directly bind the inner concave surface of GST-KPNB1. The central role of the NLS in SRF–karyopherin binding is demonstrated by our finding that mutation of the NLS abolishes all such interactions in vitro (see Figure 3).
Our study shows that, even though SRF can directly bind to KPNB1 (and to KPNA2 in the cytoplasm), such binding seems unlikely to be physiologically important in airway myocytes. Instead, formation of the ternary complex, KPNA1–KPNB1–SRF, appears to be the dominant mechanism by which SRF gains nuclear entry, as demonstrated by the import assay (Figure 5). Other investigators have used permeabilized cells to study the nuclear entry of proteins (21,22). In this procedure, digitonin permeabilized cells are washed several times to remove native cytoplasmic proteins, thus allowing investigators to provide cells with an “artificial cytoplasm” reconstituted with specifically chosen recombinant proteins. In addition, the perinuclear calcium concentration is controlled via EGTA containing transport buffer. In our studies, the nuclear accumulation of SRF-GFP after 1 minute in artificial cytoplasm (import buffer) containing KPNA1 and KPNB1 demonstrates that these proteins together are sufficient to mediate SRF nuclear entry. In contrast, our import assays demonstrate no significant nuclear accumulation of SRF-GFP when KPNB1 is the only karyopherin included in the import buffer. This is further evidence that KNPB1 alone is unable to effect substantial nuclear entry of SRF. Thus, KPNA1 is an essential component of SRF NLS-dependent nuclear import.
Our study provides evidence that SRF is able to achieve nuclear entry through an auxiliary, NLS-independent mechanism. We observed that transfected CTSM cells accumulate EGFP–mNLS-SRF in the nucleus when treated with leptomycin B, a nuclear export inhibitor (Figure 2E). This accumulation is not likely the result of an NLS-dependent import mechanism, given the lack of 35S–mNLS-SRF binding to GST-KPNA1 or KPNB1. It is conceivable that EGFP-SRF proteins are able to complex with another (undetermined) protein to gain nuclear entry. Although the precise nature of the auxiliary SRF nuclear import mechanism has not been established, it is of interest that some SRF-EGFP did enter CTSM nuclei in our import assay, even in the absence of karyopherins (Figure 5). Recent reports from Xu (15, 23) demonstrate that Smad3, Smad4, or their activated complex can bind directly to the NPC in a karyopherin-independent fashion to gain nuclear entry. Whether SRF can use a similar, non–NLS-dependent mechanism remains uncertain. Potential import mechanisms used by SRF are summarized schematically in the online supplement.
One possible limitation of our study is the use of two different cell types. The experiments examining the subcellular localization of EGFP-SRF proteins and the import assays used CTSM cells. Unfortunately, we could not accomplish immunoprecipitation of KPNA1 using purchased antibodies. Given this limitation, we overexpressed tagged SRF and karyopherin proteins in HEK293 cells to examine their potential interaction. This cell line is frequently used because of its high transfection efficiency and robust expression of transfected constructs. Nevertheless, the import assays suggest that the KPNA1–SRF interaction, demonstrated in HEK293 cells, is not cell-line specific. A functional KPNA1–KPNB1 interaction in CTSM is demonstrated by the nuclear accumulation of SRF-EGFP after the addition of these proteins to the reconstituted “cytoplasm” of permeabilized CTSM. Thus, the use of two different cells lines strengthens our conclusion that KPNA1 and KPNB1 are sufficient to facilitate the nuclear entry of SRF.
SRF is a potent transcription factor that activates the promoters of several smooth muscle–specific and immediate early genes. Our findings suggest that the formation of the ternary complex, KPNA1–KPNB1–SRF, is the primary mechanism by which SRF gains nuclear entry. Additional studies of SRF nuclear export may help determine whether its nuclear-cytoplasmic shuttling is altered in disease states such as asthma, atherosclerotic heart disease, or pulmonary fibrosis where SRF might play an important role in pathogenesis.
Supplementary Material
Acknowledgments
The authors thank Michael Malim and Karsten Weis for plasmid constructs.
Footnotes
This work was supported by NHLBI grant SCOR HL56399 (J.S.), by a postdoctoral research fellowship grant from TSANZ (D.J.F.), by The ALA-Blowitz Ridgeway Foundation (B.C.M.), and by the ATS-The LAM Foundation (B.C.M.).
This article contains an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI:10.1165/rcmb.2008-0393OCon December 3, 2010
Author Disclosure: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Camoretti-Mercado B, Liu HW, Halayko AJ, Forsythe SM, Kyle JW, Li B, Fu Y, McConville J, Kogut P, Vieira JE, et al. Physiological control of smooth muscle-specific gene expression through regulated nuclear translocation of serum response factor. J Biol Chem 2000;275:30387–30393 [DOI] [PubMed] [Google Scholar]
- 2.Liu HW, Halayko AJ, Fernandes DJ, Harmon GS, McCauley JA, Kocieniewski P, McConville J, Fu Y, Forsythe SM, Kogut P, et al. The RhoA/Rho kinase pathway regulates nuclear localization of serum response factor. Am J Respir Cell Mol Biol 2003;29:39–47 [DOI] [PubMed] [Google Scholar]
- 3.Kaplan-Albuquerque N, Garat C, Desseva C, Jones PL, Nemenoff RA. Platelet-derived growth factor-BB-mediated activation of Akt suppresses smooth muscle-specific gene expression through inhibition of mitogen-activated protein kinase and redistribution of serum response factor. J Biol Chem 2003;278:39830–39838 [DOI] [PubMed] [Google Scholar]
- 4.Ding W, Witte MM, Scott RE. Transformation blocks differentiation-induced inhibition of serum response factor interactions with serum response elements. Cancer Res 1999;59:3795–3802 [PubMed] [Google Scholar]
- 5.Gauthier-Rouviere C, Vandromme M, Lautredou N, Cai QQ, Girard F, Fernandez A, Lamb N. The serum response factor nuclear localization signal: general implications for cyclic amp-dependent protein kinase activity in control of nuclear translocation. Mol Cell Biol 1995;15:433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adam SA. The nuclear pore complex. Genome Biol 2001;2:REVIEWS0007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol 2004;14:505–514 [DOI] [PubMed] [Google Scholar]
- 8.Kobe B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat Struct Biol 1999;6:388–397 [DOI] [PubMed] [Google Scholar]
- 9.Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell 1986;46:567–574 [DOI] [PubMed] [Google Scholar]
- 10.Camoretti-Mercado B, Fernandes DJ, Dewundara S, Churchill J, Ma L, Kogut PC, McConville JF, Parmacek MS, Solway J. Inhibition of transforming growth factor beta-enhanced serum response factor-dependent transcription by Smad7. J Biol Chem 2006;281:20383–20392 [DOI] [PubMed] [Google Scholar]
- 11.Norman C, Runswick M, Pollock R, Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 1988;55:989–1003 [DOI] [PubMed] [Google Scholar]
- 12.Pellegrini L, Tan S, Richmond TJ. Structure of serum response factor core bound to DNA. Nature 1995;376:490–498 [DOI] [PubMed] [Google Scholar]
- 13.Prywes R, Zhu H. In vitro squelching of activated transcription by serum response factor: evidence for a common coactivator used by multiple transcriptional activators. Nucleic Acids Res 1992;20:513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao Z, Liu X, Lodish HF. Importin beta mediates nuclear translocation of smad 3. J Biol Chem 2000;275:23425–23428 [DOI] [PubMed] [Google Scholar]
- 15.Chen HB, Rud JG, Lin K, Xu L. Nuclear targeting of transforming growth factor-beta-activated Smad complexes. J Biol Chem 2005;280:21329–21336 [DOI] [PubMed] [Google Scholar]
- 16.Chan CK, Hubner S, Hu W, Jans DA. Mutual exclusivity of DNA binding and nuclear localization signal recognition by the yeast transcription factor gal4: implications for nonviral DNA delivery. Gene Ther 1998;5:1204–1212 [DOI] [PubMed] [Google Scholar]
- 17.Forwood JK, Lam MH, Jans DA. Nuclear import of Creb and AP-1 transcription factors requires importin-beta 1 and Ran but is independent of importin-alpha. Biochemistry 2001;40:5208–5217 [DOI] [PubMed] [Google Scholar]
- 18.Lam MH, Briggs LJ, Hu W, Martin TJ, Gillespie MT, Jans DA. Importin beta recognizes parathyroid hormone-related protein with high affinity and mediates its nuclear import in the absence of importin alpha. J Biol Chem 1999;274:7391–7398 [DOI] [PubMed] [Google Scholar]
- 19.Moore JD, Yang J, Truant R, Kornbluth S. Nuclear import of Cdk/cyclin complexes: identification of distinct mechanisms for import of Cdk2/cyclin E and Cdc2/cyclin B1. J Cell Biol 1999;144:213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem 2007;76:647–671 [DOI] [PubMed] [Google Scholar]
- 21.Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol 1990;111:807–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jankowski R, Czubryt MP, Pierce GN. The nuclear protein import assay in vascular smooth muscle cells. J Pharmacol Toxicol Methods 2000;44:421–427 [DOI] [PubMed] [Google Scholar]
- 23.Xu L, Alarcon C, Col S, Massague J. Distinct domain utilization by Smad3 and Smad4 for nucleoporin interaction and nuclear import. J Biol Chem 2003;278:42569–42577 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.