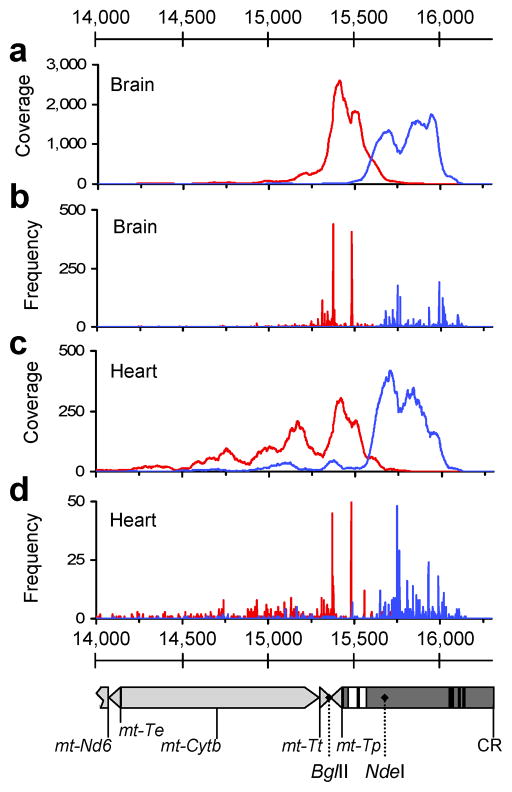
Figure 2. Localization of R-F oriented read pairs and R-F breakpoints within the CRM region in PolgD257A/D257A assemblies.
(a, c) Coverage maps of R-F oriented read pairs extracted from brain and heart assemblies from P1 (PolgD257A/D257A). Forward oriented reads are shown in blue and reverse oriented reads in red. (b, d) Frequencies of breakpoints positions in brain and heart libraries from P1. Positions of 5′ bases (F) shown in blue and 3′ bases (R), in red. Detail of mtDNA nt14,000–16,299 aligned to panels a-d is shown below. Genes (light grey) and control region (dark grey) are depicted. ETAS1 and -2 (extended termination sequence) are represented as white boxes and CSBI, -II and -III (conserved sequence blocks) as black boxes. Relevant restriction sites are labeled with dotted lines.