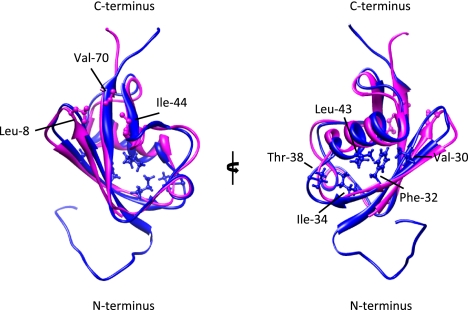
Figure 1.
UBD and SIM interaction surfaces on Ub and SUMO are not conserved. Structural alignment of a molecular ribbon representation of Ub (magenta; Protein Data Bank [PDB]: 1aar) and SUMO-3 (cyan; PDB: 2rpq). On Ub, the canonical Leu 8, Ile 44, and Val 70 residues that contact UBDs are indicated (see Dikic et al. 2009). Val 30, Phe 32, Ile 34, Thr 38, and Leu 43 on SUMO3 have been shown to contact a canonical SIM in MCAF1 (the MBD1 [methyl-CpG-binding domain protein 1]-containing chromatin-associated factor 1) (Sekiyama et al. 2008). A similar surface in SUMO1 or SUMO2/3 has also been shown to be involved in binding to the hydrophobic SIM region of PIAS family members (Hecker et al. 2006). Remarkably, positively charged residues in SUMO paralogs, including Lys 33 of SUMO2, which is conserved in the SLD2 of UAF1, contribute to SIM binding. Note that opposite surfaces on SUMO and Ub serve as interaction platforms for the respective binding modules. The image was generated using University of California at San Francisco Chimera (release date May 24, 2011; http://www.cgl.ucsf.edu/chimera).