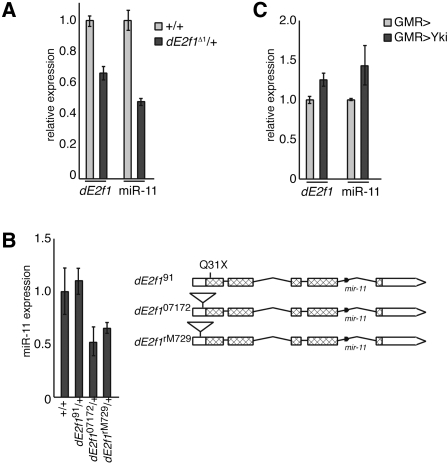
Figure 1.
Expression of miR-11 parallels expression of dE2f1. (A) cDNA was prepared from RNA extracted from Canton S (wild-type) or de2f1Δ1 adult flies. The expression of mir-11 and the de2f1 mRNA were measured using TaqMan or standard real-time PCR, respectively, and normalized to β-tubulin or RpP0 levels. Expression of normalized dE2f1 and miR-11 in wild type was designated as 1.0, and expression in de2f1Δ1 samples was compared. (B) RNA was extracted from third instar larvae. miR-11 expression in the indicated genotypes was determined and compared with wild type, following normalization to β-tubulin and RpP0 expression. A diagram of the dE2f1 exon/intron structure, mutant alleles, and mir-11 gene examined is shown. The dE2f1 ORF corresponds to hatched bars, while UTRs are indicated by white bars. Introns are represented by horizontal lines. The dE2f1rM729 P-element insertion is 48 nt upstream of the initiator methionine, the dE2f107172 P-element insertion is 33 nt upstream of the initiator methionine, and the dE2f191 allele is a C91T point mutation, giving a Q31X early translation termination codon (Mlodzik and Hiromi 1992; Duronio et al. 1995; Brook et al. 1996). (C) Flies carrying the GMR-Gal4 transgene were crossed to wild-type or UAS-ykiS168A flies. RNA was extracted from third instar larval eye discs, and miR-11, dE2f1, β-tubulin, and RpP0 expression was measured by qRT–PCR. Expression levels shown are relative to GMR-Gal4/+.