The Sir2/3/4 complex is a key component of the Sir gene silencing machinery in S. cerevisiae. This structure–function study examines the unique folding structure of the Sir3 AAA+ domain and provides insight into new interaction surfaces between Sir3 and Sir4 and into its chromatin-binding properties.
Keywords: SIR complex, HMR, HML, gene silencing, telomere, Sir2
Abstract
The silent information regulator 2/3/4 (Sir2/3/4) complex is required for gene silencing at the silent mating-type loci and at telomeres in Saccharomyces cerevisiae. Sir3 is closely related to the origin recognition complex 1 subunit and consists of an N-terminal bromo-adjacent homology (BAH) domain and a C-terminal AAA+ ATPase-like domain. Here, through a combination of structure biology and exhaustive mutagenesis, we identified unusual, silencing-specific features of the AAA+ domain of Sir3. Structural analysis of the putative nucleotide-binding pocket in this domain reveals a shallow groove that would preclude nucleotide binding. Mutation of this site has little effect on Sir3 function in vivo. In contrast, several surface regions are shown to be necessary for the Sir3 silencing function. Interestingly, the Sir3 AAA+ domain is shown here to bind chromatin in vitro in a manner sensitive to histone H3K79 methylation. Moreover, an exposed loop on the surface of this Sir3 domain is found to interact with Sir4. In summary, the unique folding of this conserved Sir3 AAA+ domain generates novel surface regions that mediate Sir3–Sir4 and Sir3–nucleosome interactions, both being required for the proper assembly of heterochromatin in living cells.
Eukaryotic DNA is packaged into nucleosomal chromatin by core histones, while further folding into higher-order structures is mediated by nonhistone proteins (Woodcock 2006). Different degrees of compaction lead to the organization of genomes into euchromatic and heterochromatic domains that are essential to maintain the gene expression programs driving development and differentiation in higher organisms (Ehrenhofer-Murray 2004).
Heterochromatin tends to be domain-specific and represses genes found adjacent to heterochromatin-inducing repetitive DNA or silencers. The budding yeast Saccharomyces cerevisiae contains three heterochromatin-like regions: silent mating-type loci HML and HMR, which regulate cell identity; subtelomeric regions; and the rDNA locus (Rusche et al. 2003). Heterochromatin at the HM loci and the telomeres is characterized by the presence of the silent information regulator (SIR) proteins Sir2, Sir3, and Sir4 (Rine and Herskowitz 1987), which are essential for silencing and form an archetypal multisubunit complex required for heterochromatin assembly and maintenance. The Sir2 component is an NAD+-dependent histone deacetylase (Imai et al. 2000; Smith et al. 2000; Tanner et al. 2000), whereas Sir3 and Sir4 are structural subunits that lack a catalytic activity, but bind histones and DNA (Rusche et al. 2003).
Silencing is established by first recruiting Sir2/Sir4 to chromatin via their interaction with proteins that bind specific DNA elements flanking the genes to be repressed (Rusche et al. 2003). Next, Sir2 deacetylates the histone H3 and H4 N-terminal tails (Rusche et al. 2002), thereby creating high-affinity binding sites for Sir3 on chromatin (Hecht et al. 1995). All three Sir proteins are then required for spreading of the complex along nucleosomes to repress nearby promoters (Hecht et al. 1996; Rusche et al. 2002).
Although there is extensive evidence for interactions among SIR proteins, Sir2/Sir4 complexes isolated from yeast contain very little Sir3 (Ghidelli et al. 2001; Hoppe et al. 2002; Rudner et al. 2005). In contrast, baculovirus-mediated coexpression allows the purification of a stable heterotrimeric Sir2/Sir3/Sir4 holocomplex, which has been successfully used for in vitro chromatin binding and low-resolution structural studies (Cubizolles et al. 2006; Martino et al. 2009; Oppikofer et al. 2011; S Kueng and SM Gasser, unpubl.). In vivo, either the holocomplex may form preferentially on chromatin, or else the association of Sir3 may be enhanced by the Sir2 deacetylation reaction (Liou et al. 2005). It was proposed that SIR complex stabilization might reflect structural changes induced by a small metabolite generated by the Sir2 deacetylase enzyme O-acetyl-ADP-ribose, or OAADPR (Tanner et al. 2000; Liou et al. 2005). Because this compound increases the affinity of the SIR complex for nucleosome binding in vitro (Martino et al. 2009), we and others have hypothesized that binding of OAADPR may be important either for the association of Sir3 with the Sir2/Sir4 subcomplex, or for Sir3–Sir3 interaction and the “spreading” of the complex along the chromatin fiber (Gasser and Cockell 2001; Liou et al. 2005; Ehrentraut et al. 2010). On the other hand, silencing can be genetically rendered at least partially independently of OAADPR production (Chou et al. 2008). Thus, the precise role of this Sir2 metabolite or other nucleotides in regulating the SIR complex is not known.
Interestingly, Sir3 shares its general protein architecture with Orc1, the large subunit of the replicative origin recognition complex (ORC), in that both contain an N-terminal bromo-adjacent homology (BAH) domain (amino acids 1–214) (Connelly et al. 2006; Hou et al. 2006; Hickman and Rusche 2010) and the C-terminal AAA+ domain (amino acids 532–845) (Supplemental Fig. S1). Typically, in the AAA+ family of proteins, the AAA+ domain binds and hydrolyzes ATP, although in the case of Sir3, key residues that are normally required for ATP binding and catalysis appear to be missing (Bell et al. 1995; Neuwald et al. 1999). On the other hand, Sir3 is known to interact with multiple factors involved in the formation of silent chromatin (for review, see Norris and Boeke 2010). Notably, it interacts with itself as well as with Sir4 (Moretti et al. 1994), histones H3 and H4 (Hecht et al. 1995), and the DNA-binding protein Rap1 (Moretti et al. 1994; Chen et al. 2011).
Interactions with histones have been attributed to the Sir3 N-terminal BAH domain (Onishi et al. 2007; Buchberger et al. 2008), which shares a higher degree of conservation with the Orc1 BAH domain (50% identity/65% similarity) than do the respective AAA+ domains (27% identity/43% similarity). Indeed, a swapping of BAH domains between Sir3 and Orc1 proteins generates functional chimeras, while a similar exchange between AAA+ domains does not (Bell et al. 1995). Mutations in the BAH domain region suppress silencing defects of mutations at H4K16 and H3K79 in vivo (Johnson et al. 1990; Thompson et al. 2003), suggesting relevant contacts both with the H4 N terminus and on the face of the nucleosome. Importantly, methylation of H3K79 by Dot1 (van Leeuwen et al. 2002) has been argued to reduce interaction with the recombinant BAH domain (Onishi et al. 2007), but also to reduce binding to recombinant Sir3 lacking its N terminus (amino acids 620–978) (Altaf et al. 2007). Thus, it remained unresolved whether one or both Sir3 domains bind the face of the nucleosome in a methylation-sensitive manner. Intriguingly, earlier experiments indicated that parts of the Sir3 AAA+ domain (C-terminal histone-binding domain 1 [CHB1] [amino acids 623–762] and CHB2 [amino acids 799–910]) can bind histones H3 and H4 peptides in vitro (Hecht et al. 1995) and that a Sir3 fragment (amino acids 620–978) could compete with Dot1 for binding to the H4 tail, blocking H3K79 methylation (Altaf et al. 2007). Given that this same domain overlaps with the minimal region that mediates binding between Sir3 and Sir4 (amino acids 464–728) (King et al. 2006), it remained unclear which of these activities attributed to the AAA+ domain was crucial for SIR-mediated repression.
In this study, we sought to dissect the function of the Sir3 AAA+ domain in HM and telomeric silencing by combined genetic, biochemical, and structural dissection of this ATPase-like domain. To date, only two alleles defective in Sir3 function have been isolated within its AAA+ domain (sir3-S813F and sir3-L738P) (Stone et al. 2000; Buchberger et al. 2008). To map these and other mutations generated here to structural domains, we determined the crystal structure of the Sir3 AAA+ domain (amino acids 530–845). This revealed several novel, silencing-specific features within the Sir3 AAA+ domain. First, we identified a loop on the AAA+ domain surface comprising residues 657–659 as the critical site on Sir3 for interaction with Sir4. Second, we show that the core Sir3 AAA+ domain is able to bind nucleosomes in vitro in a manner that is sensitive to H3K79 methylation. Third, we identify and characterize several other surface patches that are necessary for Sir3 silencing function in vivo. Finally, we show that the Sir3 AAA+ domain has several unusual features that, surprisingly, would disfavor interaction with nucleotides. Taken together, our analysis provides a comprehensive view of the involvement of the novel folds within the Sir3 AAA+ domain that mediate protein–protein interactions crucial for the proper assembly of a functional SIR complex on unmethylated nucleosomes, a prerequisite for silent chromatin formation in vivo.
Results
The Sir3 AAA+ domain evolved a noncanonical protein function and forms a repeating, oligomeric assembly within the crystal
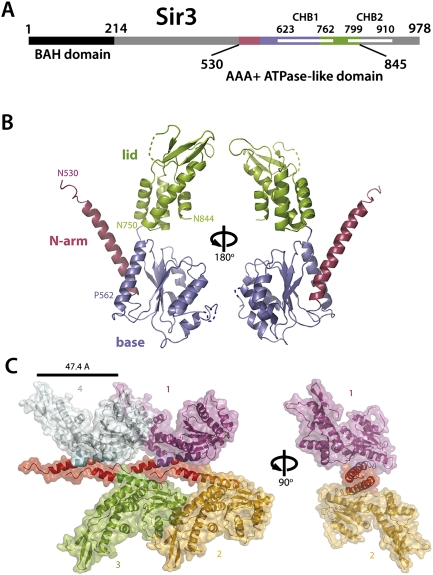
The Sir3 protein arose through a gene duplication of Orc1, with which it shares an N-terminal BAH domain and a C-terminal AAA+ ATPase-like domain (Fig. 1A; Norris and Boeke 2010). While the function of the BAH domain has been studied extensively, there is only limited knowledge available on the AAA+ domain of Sir3 (Bell et al. 1995; Hickman and Rusche 2010). Given that the Orc1 AAA+ domain cannot substitute for the Sir3 equivalent, it is assumed that the Sir3 C-terminal domain (CTD) has evolved additional functions that are crucial for transcriptional gene silencing, but has lost functions necessary for replication initiation. To gain information about the AAA+ domain of Sir3, we determined its structure. Diffracting crystals of Sir3 (amino acids 530–845) were obtained, and the structure containing two copies per asymmetric unit was solved using the single-wavelength anomalous diffraction (SAD) method and refined to 2.8 Å resolution (Fig. 1B; Supplemental Table I).
Figure 1.
The Sir3 AAA+ ATPase-like domain is a structurally divergent family member. (A) Schematic representation of the Sir3 domain structure. (B) Ribbon representation of the Sir3 AAA+ crystal structure spanning amino acids 530–844. The structure is shown in two orientations, rotated through 180°, and colored as in the schematic. Disordered or absent residues are indicated by dashed lines. The relevant subdomains are depicted in red (N-arm), blue (base), and green (lid). Residues marking subdomain boundaries are also indicated. (C) Crystal lattice contacts involving the N-arm helices of four crystallographically related molecules (differently colored, labeled 1–4). The lattice arrangement is depicted in two orientations. Molecules 3 and 4 are omitted for clarity in the right orientation.
The Sir3 AAA+ domain clearly shows the expected canonical AAA+-like architecture, consisting of the characteristic “base” subdomain (Rossman fold), as well as the “lid” subdomain containing a four-helix bundle. However, the crystal structure also revealed several unusual features in the Sir3 AAA+ fold (Fig. 1B), including an N-terminal α-helical extension and internal conformational differences distinct from the related Orc1 domain.
The first unusual feature of this AAA+ domain is an elongated N-terminal α-helical arm that protrudes from the base (referred to as the “N-arm”), which in the context of the crystal acts as a platform for oligomerization (Fig. 1C). Specifically, these N-terminal α helices are involved in an unusual staggered lattice contact forming a continuous, oligomeric coiled-coil structure. The minimal repeating unit of this unique lattice contact is formed by three crystallographically related Sir3 molecules. The combined, predominantly hydrophobic interfaces between the three crystallographically related molecules (e.g., 1–3) bury ∼810 Å2 of molecular surface (PISA server) (Krissinel and Henrick 2007).
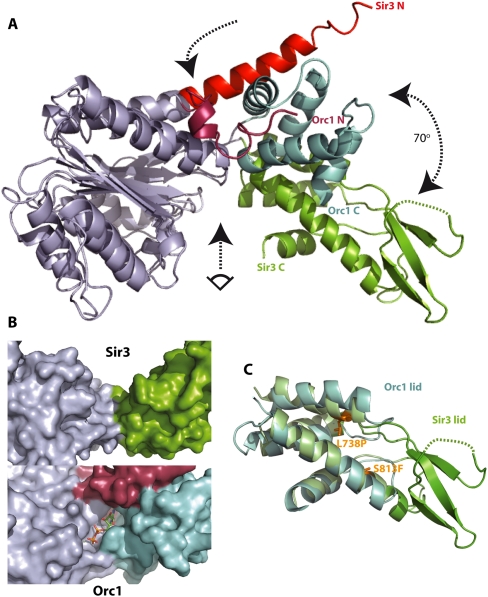
A second unusual feature of the AAA+ architecture of Sir3 became evident from the structural superimposition of Sir3 with its closest structural homolog, Sulfolobus solfataricus ORC1/cdc6 (2QBY,A) (Fig. 2A,C; Dueber et al. 2007). Both subdomain folds (base and lid) of Sir3 superimpose well with the corresponding canonical folds present in ORC1/cdc6, but their relative orientation differs remarkably. Superimposition of the base subdomains (1.6 Å root mean square deviation [RMSD] between corresponding 96 Cα atoms) nicely highlights the structural differences between the two structures (Fig. 2A; Supplemental Fig. S1). Notably, the lid domains in the two structures are rotated by ∼70° around a pivot point at the “hinge” region between the two subdomains, creating a more “open” overall conformation in Sir3. The N-terminal residues in Sir3 form the N-arm structure, as described above, whereas in ORC1/cdc6 the corresponding N-terminal residues intrude between the base and lid subdomains and form a substantial part of the nucleotide-binding pocket in ORC1/cdc6, as well as stabilize the overall conformation of the structure (Fig. 2B, bottom). The conformational differences in Sir3 preclude the formation of a classical nucleotide-binding pocket and instead form a wide and shallow groove (Fig. 2B, top). This, combined with the fact that the classical P-loop motif (GXXXXGK[S/T]) present in ORC1/Cdc6 is not present in Sir3 (amino acids 578–583), argues that the AAA+ domain of Sir3 has lost nucleotide-binding capability. Indeed, attempts to detect the association of ATP or ADP-ribose with the recombinant AAA+ domain by isothermal titration calorimetry assays were not successful (Supplemental Fig. S2). Finally, the superimposition of the four-helix bundle in the “lid” region (2.1 Å RMSD between 34 helical Cα atoms) indicated an extension of the loop linking α helices 1 and 2 in this subdomain of Sir3 by a threefold anti-parallel β sheet and an unstructured loop (Fig. 2C), which raises the possibility that it would create a novel functional domain. In summary, comparison of the structure of the Sir3 AAA+ domain with other AAA+ ATPase-like proteins reveals several features unique to Sir3, the most striking of which is the loss of the nucleotide-binding pocket.
Figure 2.
The Sir3 AAA+ ATPase-like domain has a strikingly different conformation from its closest structural relative, Orc1/cdc6, and does not contain a nucleotide-binding pocket. (A) Superposition of the AAA+-like domains of Sir3 and of S. solfataricus ORC1/cdc6 (2qby, A). Structural alignments were produced by superposition of the residues within the base subdomain of the two structures (RMSD of Cα atoms < 1.6 Å). For clarity, base subdomains are both colored gray. Other subdomain features are colored differently to highlight differences. The arrows indicate the hinge region (top arrow), the rotation between the lid domains of Sir3 and ORC1/Cdc6 (right arrow), and the view into the (potential) nucleotide-binding pocket shown in B (bottom arrow). (B) Surface representation of the presumed nucleotide-binding pocket of Sir3 (top) compared with the ADP bound nucleotide-binding pocket in S. solfataricus ORC1 (bottom). The shallow and wide groove in Sir3 is not compatible with a suggested OAADPR-binding function. (C) Superposition of the lid subdomains of Sir3 and of S. solfataricus ORC1 (2qby, A). Structural alignments were produced by superposition of the helical residues within the lid subdomains of the two structures (RMSD of Cα atoms = 2.1 Å). Additional structural features in the Sir3 lid subdomain are evident (green). These include a threefold anti-parallel β sheet containing mostly positively charged residues. The location of two known point mutants causing a sir3 phenotype (Stone et al. 2000; Buchberger et al. 2008) are also indicated.
Chromatin binding of the Sir3 AAA+ domain is sensitive to H3K79 methylation
Full-length Sir3 has the ability to bind to chromatin (Georgel et al. 2001), and this binding is sensitive to methylation in the nucleosome core region at H3K79 (Martino et al. 2009; Oppikofer et al. 2011). As discussed above, recognition of the nucleosomal face has been attributed to both the Sir3 N-terminal BAH domain as well as a large C-terminal fragment (amino acids 620–978) that encompasses the AAA+ domain (Hecht et al. 1995; Connelly et al. 2006; Altaf et al. 2007; Onishi et al. 2007). Genetic and biochemical evidence indicates that the BAH domain is sensitive to H3K79 methylation (Onishi et al. 2007; Buchberger et al. 2008; Sampath et al. 2009), yet the binding of the large C-terminal region of Sir3 to an H3 peptide (amino acids 67–89) is also sensitive to methylation on K79 (Altaf et al. 2007).
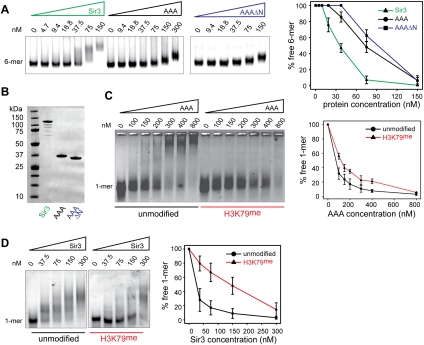
To address the contribution of the AAA+ domain to chromatin binding, we compared its association with recombinant nucleosomal arrays with that of full-length Sir3. Increasing amounts of recombinant full-length Sir3 and AAA+ domain were incubated with a constant amount of a 6-mer of regularly spaced nucleosomes reconstituted on the 601-Widom sequence, as described earlier (Martino et al. 2009). Binding was analyzed by native agarose gel electrophoresis (Fig. 3A). We note that both the AAA+ and, to a large extent, full-length Sir3 are monomers in solution under the conditions used here (Supplemental Fig. S3; Cubizolles et al. 2006), although under lower-salt conditions, Sir3 is able to oligomerize in vitro (Liou et al. 2005; McBryant et al. 2006). We also included a shortened AAA+ domain lacking the N-arm (AAAΔN, amino acids 545–845), since the absence of these residues in full-length Sir3 caused a significant loss of telomeric silencing in vivo (see below, sir3-1073) (Fig. 3C). The titrations show that the AAA+ domain has chromatin-binding capacity, although it is twofold to threefold reduced compared with full-length Sir3 (Fig. 3A). The AAAΔN protein bound chromatin like the full-length AAA+ domain, indicating that this N-arm is not a major chromatin interaction site.
Figure 3.
Binding of the Sir3 AAA+ ATPase-like domain to chromatin was sensitive to methylation of H3K79. (A) The Sir3 protein, the Sir3 AAA+ ATPase-like domain (AAA; amino acids 530–845), or an N-terminal truncation (AAAΔN; amino acids 545–845) was titrated over a constant amount (25 nM) of unmodified 6-mer nucleosomes. (B) SDS-PAGE gel of 1 μg of the Sir3 protein, the Sir3 AAA+ domain, and the N-terminal truncation used in the experiments above (staining with Coomassie brilliant blue). (C) The Sir3 AAA+ ATPase-like domain was titrated over a constant amount (25 nM) of unmodified or H3K79me Cy3-147 mononucleosomes. Samples were separated by native agarose gel electrophoresis, and Cy3-labeled DNA was visualized. The images are representative of at least three independent experiments, and quantifications show the mean value ± SEM of the percent of unbound chromatin compared with the input. (D) Full-length Sir3 was titrated over a constant amount (25 nM) of unmodified or H3K79me mononucleosomes; three independent experiments were analyzed and plotted as described in C.
To see whether the binding of the AAA+ domain was sensitive to H3K79 methylation in the context of nucleosomes, we incubated the AAA+ domain with mononucleosomes that had been methylated by recombinant yeast Dot1 (Fig. 3C). These nucleosomes are methylated between 50% and 70%, containing mono-, di-, and trimethylated K79 residues (Martino et al. 2009). Previous studies showed that all methylated forms are functional in disrupting silencing (Frederiks et al. 2008). Indeed, the binding affinity of the AAA+ domain to nucleosomes was reduced by roughly twofold upon H3K79 methylation. In the same conditions, the binding affinity of full-length Sir3 was reduced by approximately sixfold by H3K79 methylation (Fig. 3D). We conclude that the Sir3 AAA+ domain binds both a hexamer and a mononucleosome, with a preference for unmethylated H3K79, yet it is likely that the N-terminal BAH domain also contributes to the pronounced sensitivity of full-length Sir3 to H3K79 methylation.
Mutant alleles reveal a functional requirement for the Sir3 AAA+ domain
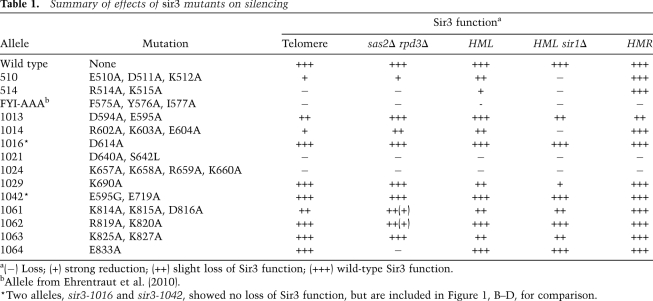
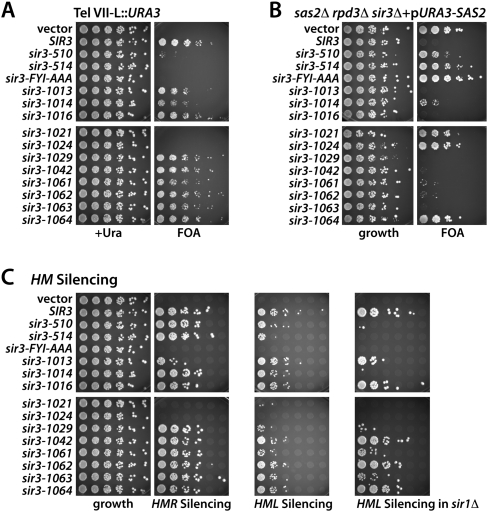
To obtain an integrated view of functionally relevant residues in the Sir3 AAA+ domain, we performed a systematic mutational analysis by mutating to alanine every charged residue (D, E, R, and K) between Sir3 residues 532 and 834 and tested how each substitution affects the silencing function of full-length Sir3 (Table 1; Supplemental Table II). Additionally, we tested 10 mutant alleles in which we altered charged residues between amino acids 407 and 523, a region upstream of the AAA+ domain (Supplemental Table I). Because neighboring D, E, R, or K residues were combined into single alleles, we generated 74 novel sir3 alleles, which were subjected to functional analysis. All of the mutant proteins generated were expressed at the same level as wild-type Sir3, ruling out that changes were due to altered stability of the mutant allele (Supplemental Fig. S4). We tested silencing by scoring the ability of the sir3 allele to support the repression of a subtelomeric URA3 reporter (Gottschling et al. 1990). This assay revealed nine new sir3 alleles within the AAA+ domain that reduce the level of Sir3-mediated telomeric silencing, as scored by colony growth on 5-FOA plates (Fig. 4A; Table 1; see Supplemental Fig. S1 for location of mutants on Sir3–Orc1 sequence alignment). Analysis of the more N-terminal alleles also identified two novel mutations (sir3-510 and sir3-514) that severely weakened Sir3 function (Fig. 4A; Table 1). We conclude that the Sir3 AAA+ domain does indeed contain multiple domains crucial for the function of Sir3 in telomeric repression.
Table 1.
Summary of effects of sir3 mutants on silencing
Figure 4.
Mutational analysis of the Sir3 AAA+ domain. (A) Telomeric silencing assay with selected sir3 alleles generated in the alanine scan. The SIR3 alleles were introduced into a TEL-VII-L::URA3 sir3Δ strain and tested for their ability to silence the subtelomeric URA3 reporter by plating serial dilutions on 5-FOA plates. Strains were grown for 2 d (5-FOA three) at 30°C. (B) Test of the selected sir3 alleles for their ability to restore the sas2Δ rpd3Δ synthetic lethality. sir3 alleles were introduced into sas2Δ rpd3Δ sir3Δ carrying SAS2 on a URA3-marked plasmid. Strains were serially diluted and tested for their ability to lose the pURA3-SAS2 plasmid on 5-FOA plates. (C) Test of sir3 alleles for their activity in HM silencing. sir3 alleles were introduced into a MATα sir3Δ strain to test for HMR silencing, and into MATa sir3Δ and MATa sir1Δ sir3Δ cells to test for HML silencing. For mating, each dilution of the strains was mixed with 0.3 OD of a mating tester strain of the opposite mating type, spotted on minimal medium for diploid selection, and incubated for 2 d at 30°C. Growth assays for the HML silencing strains showed equal growth of all strains, but were omitted for clarity.
While silent chromatin is generally hypoacetylated at all known histone residues (Braunstein et al. 1993), the deacetylation of histone H4K16 is particularly critical for both silencing per se (Johnson et al. 1990; Megee et al. 1990; Park and Szostak 1990) and the spreading of the SIR complex along the chromatin fiber (Hecht et al. 1996). As a second silencing assay, we took advantage of the fact that compromised Sir3 function suppresses the synthetic lethality of a double mutant lacking both the H4K16 histone acetyltransferase Sas2 and the histone deacetylase Rpd3 (Ehrentraut et al. 2010). We presume that the sas2Δ rpd3Δ lethality is caused by the SIR complex spreading into subtelomeric regions, where it represses an essential subtelomeric gene, because a reduction of Sir3 activity can be scored as restored growth of the sas2Δ rpd3Δ mutant. We thus introduced the sir3 alleles into a strain lacking Sas2 and Rpd3 and scored for colony growth. Generally, this assay recapitulated the results of the TelVIIL::URA3 silencing test (Fig. 4B; Table 1). Both assays confirmed telomeric silencing defects in 11 new sir3 alleles, with effects ranging from slight to complete loss of repression. Interestingly, we found one allele (sir3-E833A/sir3-1064) that completely rescued the sas2Δ rpd3Δ lethality (arguing for a loss of silencing capacity), although it showed no TelVIIL::URA3 silencing defect (Fig. 4A,B). This may indicate a differential effect on silencing of the presumed essential subtelomeric gene, as opposed to the artificial URA3 reporter.
Since the Sir3 protein is also essential for HM silencing, we next tested the effect of the sir3 alleles on HMR and HML silencing. HML silencing was tested in the context of sir1Δ, which allows detection of more subtle silencing defects (Stone et al. 2000). Significantly, the sir3 alleles that showed the strongest telomeric derepression effects (sir3-1021, sir3-1024, sir3-514, and FYI-AAA) also abolished HM silencing at HMR and/or HML (Fig. 4C). Furthermore, sir3 alleles that showed an intermediate telomeric silencing defect differed in their effects on HM silencing. For instance, the sir3-1014 allele that revealed a moderate telomeric silencing defect showed derepression at HML, which was further enhanced by sir1Δ (Fig. 4C), but this allele showed no silencing defect at HMR. Interestingly, the sir3-1013 allele showed a slight HMR silencing defect without displaying telomeric silencing defects (Fig. 4C).
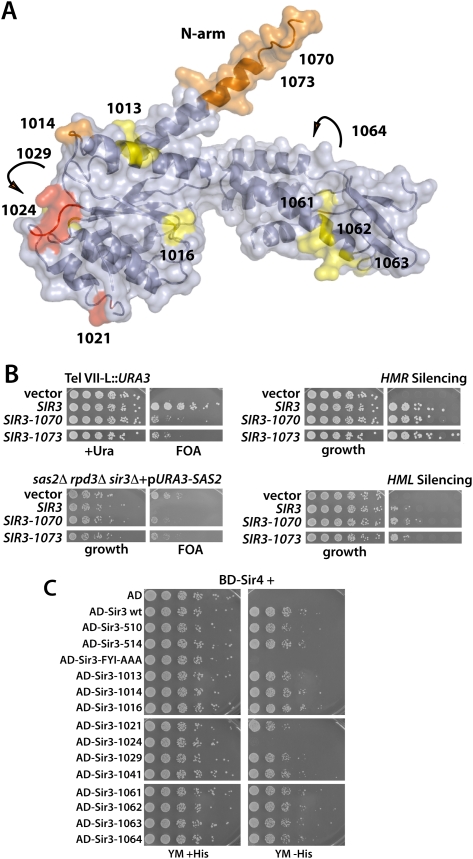
Since the crystal structure of the Sir3 AAA+ domain revealed an unusual N-arm (Fig. 1B), we sought to determine the in vivo relevance of this feature for Sir3 silencing function. To test this, we generated two mutants that either replaced the first 12 amino acids within this arm by an alanine residue (sir3-1070; amino acids 532–543 replaced) or completely deleted 17 residues (sir3-1073; Δ530–546) (Fig. 5A). Both resulting alleles displayed a significant loss of telomeric silencing when scored as derepression of subtelomeric URA3 or as a loss of sas2Δ rpd3Δ lethality (Fig. 5B). Conversely, these alleles did not affect HM silencing (Fig. 5B). Thus, the alleles affecting the N-arm showed a specific loss of Sir3 function at telomeres.
Figure 5.
Mapping mutant alleles on the Sir3 AAA+ structure. (A) Surface representation of the crystal structure of the Sir3 AAA+-like domain. The positions of the sir3 alleles listed in Table 1 are indicated on the surface in red (strong phenotype), orange (intermediate phenotype), or yellow (weak phenotype). Arrows indicate that the alleles sir3-1029 and sir3-1064 are located on the reverse side of Sir3 in this view. (B) Mutational analysis of the N-terminal arm of Sir3 observed in the crystal structure. Two mutations that partially deleted the N-terminal arm sequence (D amino acids 530–546; sir3-1073) or mutated every amino acid within this region to alanine (sir3-1070) were tested for their effect on telomeric and HM silencing as in Figure 4. (C) The alleles that affected Sir3 function were tested for Sir3–Sir3 and Sir3–Sir4 interaction by a two-hybrid assay. Strains (AEY3055 transformed with the respective plasmids) were tested for activation of the two-hybrid reporter HIS3 by plating serial dilutions on minimal medium with or without histidine.
In summary, mutational analysis of the Sir3 AAA+ domain identified 13 new alleles that either completely abolished Sir3 function or revealed partial silencing defects, although none of the alleles were dominant in the telomeric URA3 silencing assay (data not shown). Of note, none of the mutations generated in the Sir3-specific extension of the “lid” region (sir3-1048 to sir3-1055) (Supplemental Table I) caused a silencing defect. Thus, it remains unclear whether or how this structural feature contributes to Sir3 function. In summary, this extensive mutagenesis confirms the crucial role played by the Sir3 AAA+ domain in SIR-mediated heterochromatin formation.
Mapping of allelic mutants onto the structure identifies protein interaction surfaces
To more closely characterize the regions of the Sir3 AAA+ domain identified as important for silencing, we mapped the position of our sir3 alleles onto the crystal structure (Fig. 5A). Significantly, all mutations from this study that affected Sir3 function map close to the surface of our Sir3 structure, suggesting that the phenotypic effects are most likely due to the loss of interactions with other silencing factors, rather than resulting from loss of the three-dimensional structure of the AAA+ fold.
Formation of heterochromatin through the SIR complex requires multiple interactions between the SIR complex and other proteins, as well as multivalent interactions among SIR subunits (Rusche et al. 2003). We therefore tested the interaction between Sir3 or sir3 alleles (amino acids 307–978) with Sir3 (amino acids 307–978) and Sir4 (amino acids 839–1358) using a two-hybrid assay (Moretti et al. 1994). In a previous study, we found that the sir3 FYI-AAA allele (sir3-F575A, Y576A, I577A) completely abolished Sir3–Sir3 and Sir3–Sir4 interactions (Ehrentraut et al. 2010), yet the FYI-AAA allele is located in the core of the “base” subdomain on β sheet 1 (see Supplemental Fig. S1) and is predicted to affect the overall fold of Sir3. In our new analysis of Sir3–Sir4 and Sir3–Sir3 interaction, we identified an allele located on the surface of Sir3 (sir3-1024, sir3-K657A, K658A, R659A, K660A) that clearly abolishes Sir3–Sir4 interaction (Fig. 5C). This suggests that the exposed loop spanning Sir3 amino acids 657–660 mediates interaction between Sir3 and Sir4. Importantly, no other alleles were seen to affect this interaction (Fig. 5C; data not shown). We tested Sir3 dimerization in these alleles by two-hybrid using Sir3 fragments that contain the CTD (amino acids 843–978), a domain that is reported to be the key for Sir3 dimerization (Liaw and Lustig 2006). None of the alleles interfered with Sir3 dimerization (data not shown), probably reflecting the more efficient dimerization mediated by the extreme C terminus of Sir3. In sum, the observed loss of Sir3–Sir4 interaction for an allele mutated in amino acids 657–660 argues strongly that this cluster of residues serves as a contact site between Sir3 and Sir4. The loss of silencing in this allele indicates that this domain is likely to be necessary for the functional assembly of silent chromatin.
The Sir3–Sir4 interaction is mediated by Sir3 residues K657, K658, and R659
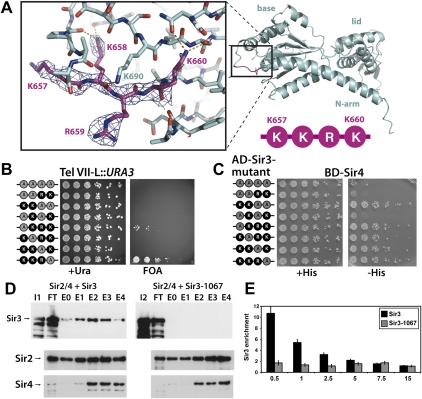
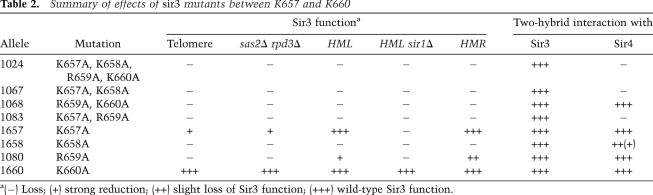
The Sir3 residues 657–660 are part of an extended loop in the base subdomain that connects α helix 4 to strand 3 of the central parallel β sheet. Residues K657 and R659 are surface-exposed, while the side chains of K658 and K660 face inward, forming part of an extensive net of hydrogen bonds that stabilizes the protein conformation in this region (Fig. 6A). Since the mutant sir3-1024 allele spanned four amino acids, we sought to separate this mutant into a series of more subtle point mutations. To this end, we divided the mutagenesis into two double mutations, four single mutations, or combinations thereof (Fig. 6B). Six of the seven resulting alleles derepressed telomeric URA3, with the K657A allele showing an intermediate effect, whereas the K660A allele showed no telomeric silencing defect (Fig. 6B; Table 2). Here, also, the measurement of telomeric silencing through restoration of the sas2Δ rpd3Δ lethality reflected the Tel VII-L::URA3 assay (Supplemental Fig. S5; Table 2). Furthermore, the double mutations and the K658A mutation also led to a loss of HM silencing at both HM loci (Supplemental Fig. S5; Table 2). Since even single amino acid substitutions in this region led to a pronounced loss of Sir3 function, our analysis identifies a strong relevance of amino acids 657–659 in SIR function.
Figure 6.
Detailed mutational analysis of the Sir3 residues 657–660. (A) Representation of the Sir3 region spanning the residues K657, K658, R659, and K660. The loop residues 657–660 are shown in stick representation with the respective 2Fo − Fc electron density map contoured at 1.5 σ. Hydrogen bonds formed by the residues are indicated by orange dashed lines. (B) Telomeric silencing assay with the indicated combinations of mutations. Unmodified residues are shown in black, and positions mutated to alanine are shown in gray. The alleles were introduced into a Tel VII-L::URA3 sir3Δ strain and tested for their ability to silence the subtelomeric URA3 reporter as in Figure 4A. (C) Test of the selected sir3 alleles for Sir3–Sir4 two-hybrid interaction as in Figure 5B. (D) In vitro SIR complex assembly with Sir3 and Sir3-1067. Cells infected with Sir2/4, Sir3, or Sir3-1067 expression constructs were lysed, and extracts were mixed. Total extract (I1: extract Sir3; I2: extract Sir3-1067), flow-through (FT), and the eluted fractions were analyzed by Western blotting against HA-tagged Sir3 (top panel), Sir2 (middle panel), and Sir4 (anti-CBP) (bottom panel). (E) Sir3-1067 was unable to bind to telomeres. Sir3 binding at the right telomere of chromosome VI is shown as enrichment in ChIP experiments relative to the enrichment at the control gene SPS2. The amount of enrichment is given as a function of the distance to the telomere end in kilobases. ChIPs were performed with antibodies against HA-Sir3. Error bars give standard deviations.
Table 2.
Summary of effects of sir3 mutants between K657 and K660
We next asked whether the dissection into more subtle mutations also disrupted the Sir3–Sir4 interaction. Significantly, abrogation of the interaction required mutation of at least two residues at positions 657, 658, or 659 (Fig. 6C; Table 2). Whereas none of these alleles lost Sir3–Sir3 dimerization activity (data not shown), mutation of K657 and K658 had a significantly stronger effect on the Sir3–Sir4 interaction. This argues that Sir3 functionality relies primarily on K658, but also on K657 and R659. The requirement of K658 could indicate that an intact loop surface conformation is necessary for Sir4 interaction and that other residues in the vicinity may contribute to the interaction interface. Of note, K690 (sir3-1029), which shows a weak loss of silencing phenotype (Fig. 4C) but no loss of Sir4 binding (data not shown), is also in close vicinity to this loop and may contribute to its conformation (Fig. 6A). In sum, our mutagenesis and functional data identify an important role for this novel Sir3 surface-exposed loop region for Sir4 interaction and SIR silencing function.
Mutation of the Sir3 residues K657 and K658 disrupted SIR complex assembly in vitro
In order to directly test whether mutagenesis of the Sir4-interacting loop region in Sir3 interferes with the assembly of a native SIR holocomplex, we tested baculovirally expressed Sir3 and Sir3-1067 (Sir3-K657A, K658A) for in vitro complex assembly with Sir2 and Sir4, which bears a C-terminal calmodulin-binding domain (CBP) (Cubizolles et al. 2006). Extracts from insect cells overexpressing the Sir2/Sir4 complex were mixed with extracts from cells expressing either HA-tagged Sir3 or Sir3-1067. Interaction of the Sir2/Sir4 complex with Sir3 was tested by recovering Sir4 from the mixture with a calmodulin affinity resin, and coelution of Sir2 and Sir3 was tested by Western blotting (Fig. 6D). Significantly, wild-type Sir3 coeluted with Sir2 and Sir4 from the calmodulin column, whereas this association was completely abrogated by mutation of K657 and K658 to alanine in the sir3-1067 allele (Fig. 6D). This indicates that mutation of these residues disrupted in vitro assembly of the holo SIR complex.
A loss of interaction between Sir3 and Sir4 suggested that Sir3-1067 might not be recruited in vivo to chromatin. To test this, we measured the association of Sir3-1067 to telomeric sequences by chromatin immunoprecipitation (ChIP) (Ehrentraut et al. 2010). While wild-type Sir3 was readily detectable at subtelomeric sequences, the association of Sir3-1067 was completely abrogated (Fig. 6E), showing that mutation of the Sir3–Sir4 interaction surfaces also disrupted the recruitment of Sir3 to chromatin. This further underscored the importance of this patch of Sir3 in Sir3–Sir4 interaction and in SIR complex assembly in vitro and in vivo.
Discussion
The Sir3 protein plays a critical role in heterochromatin formation in S. cerevisiae (for review, see Norris and Boeke 2010), yet there is little mechanistic or structural insight into how Sir3 promotes the assembly of silent chromatin. While the Sir3 N-terminal BAH domain has been extensively studied both structurally (Connelly et al. 2006; Hou et al. 2006) and by mutational analysis (Stone et al. 2000; Buchberger et al. 2008; Norris et al. 2008; Sampath et al. 2009), structural information about other parts of Sir3 had not been available.
Here, we determined the crystal structure of the C-terminal AAA+ ATPase-like domain of Sir3 and were able to relate various features of this structure to the functional requirements for the in vivo function of Sir3. Foremost, we pinpointed the existence of a surface loop consisting of amino acids 657–659 as an interaction interface with Sir4. Although the Sir4 fold involved in this interaction has not been characterized, our Sir3 AAA+ structure provides the first insight into the molecular detail on the side of Sir3 for an interaction that is crucial for SIR-mediated repression. We show that mutations in this region in the context of full-length Sir3 abrogated its ability to be incorporated into a stable SIR complex in vitro and to bind to telomeres and function in silencing in vivo. Thus, these mutations likely reflect a defect in the initial recruitment of Sir3 to the chromatin-bound Sir2/Sir4 complex.
Determination of the Sir3 AAA+ structure has further allowed us to identify unexpected structural features that distinguish this domain from other members of its class of AAA+ family of ATPases. This divergence undoubtedly reflects its specialization as a silencing protein after the ORC1 gene duplication. The homology of Sir3 to AAA+-like ATPases (Neuwald et al. 1999) has prompted speculation that it may bind nucleotides, such as the Sir2-generated metabolite OAADPR (Cockell et al. 1995; Martino et al. 2009; Ehrentraut et al. 2010), yet our structural analysis argues that this is unlikely. Although Sir3 has two subdomain folds (“base” and “lid”) that individually superimpose well with those of its homolog, ORC1/Cdc6, the lid domains of the two structures are rotated by ∼70° around a hinge region in Sir3. Furthermore, Sir3 shows an N-terminal α-helical extension (“N-arm”) that, unlike the situation in ORC1/Cdc6, does not participate in the formation of a nucleotide-binding pocket. These Sir3-specific features have drastic consequences for the structure of the putative nucleotide-binding pocket in that it presents a shallow groove formed by the two subdomains, rather than a narrow binding pocket (Fig. 2B). Given this, and the absence of a classical P-loop region in Sir3 (Bell et al. 1995), we conclude that the Sir3 AAA+ domain is unlikely to bind OAADPR. This was substantiated by our inability to detect interaction between the Sir3 AAA+ domain and ADP-ribose or ATP in isothermal titration calorimetry assays (Supplemental Fig. S2). It is still possible that full-length Sir3 might form a closed pocket in the presence of the nucleotide, or that a major conformational change occurs upon binding to Sir2 and Sir4, which might narrow the groove to allow the stable retention of the Sir2 metabolite OAADPR or allow binding at another site. However, we note that multiple mutations in this region of Sir3 did not provoke a loss of silencing (Supplemental Table II). Further biochemical studies on the holocomplex will be required to conclusively determine how OAADPR affects SIR complex structure and function, and whether it involves the Sir3 AAA+ ATPase domain, either directly or indirectly.
Our biochemical analysis of the Sir3 AAA+ domain further showed that this part of Sir3 is capable of binding nucleosomes, and that it does so in a manner sensitive to methylation of H3K79. Both its affinity for nucleosomes and its sensitivity to methylation are less than that detected for full-length Sir3, since the full-length protein binds chromatin via both the BAH and the AAA+ domains. It will be interesting to determine more precisely which residues within the AAA+ domain contact the nucleosome core region and also which contact the histone N-terminal tails. It is not clear whether both domains within a single Sir3 molecule can contact the nucleosome, nor is it clear whether more than one nucleosome or nucleosomal domain would be involved in the interaction. In this respect, our mutational analysis has identified the region of the Sir3 AAA+ domain near amino acids 640–642 (sir3-1021) and, to a lesser extent, amino acids 814–816 and amino acids 825–827 (sir3-1061 and sir3-1063) as necessary for the Sir3 silencing function in yeast. Both of these regions coincide with CHB1 (623–762) and CHB2 (799–910), shown to interact with H3 and H4 N-terminal tails (Hecht et al. 1995). Given that mutations in these regions did not abrogate interaction between Sir3 and Sir4, we speculate that they could constitute the contact points of Sir3 with chromatin. Our mutational analysis further identified residues around amino acids 510–515 of Sir3, which lie outside of the AAA+ domain, as well as amino acids 602–604, which are on the domain surface, as being important for Sir3 silencing function, yet mutations in these regions did not abrogate either Sir3–Sir4 or Sir3–Sir3 interactions. Since they lie outside of the CHB domains but within a fragment of Sir3 (503–970) that interacts with full-length Rap1 (Luo et al. 2002), we speculate that they may contribute to Rap1 binding, or possibly contact other interaction partners.
In summary, our analysis of structure–function relationships of the Sir3 AAA+ domain has revealed a complex architecture of interaction regions scattered across the surface of the AAA+ domain. In particular, we identified a key loop region in the AAA+ that mediates Sir4 interaction, revealing one important element of the tightly regulated assembly process through which SIR complex proteins assemble and spread on chromatin to silence gene expression. Future studies will investigate the molecular mechanism, including interaction partners, for other regions whose mutation phenotypically disrupts SIR complex function, helping us to obtain an integrated view of the molecular contacts that Sir3 and other SIR complex subunits establish to form heterochromatin.
Materials and methods
Yeast strains and plasmids
The yeast strains and plasmids used in this study are listed in Supplemental Tables III, IV, and V. Yeast was grown and manipulated according to standard procedures (Sherman 1991). Yeast was grown on selective minimal plates (YM), and plates containing 5-fluoro-orotic acid (US Biological) were used to select against URA3. The HA-tagged versions of SIR3 were constructed as described (Zachariae et al. 1998).
Plasmid-borne sir3 alleles were generated using the plasmid gap repair technique in yeast. Fragments of the SIR3 gene carrying the respective mutations were generated by PCR sewing, and were introduced into linearized plasmids by homologous recombination in yeast. Mutant plasmids were amplified in Escherichia coli, and mutations were verified by sequence analysis. Derivatives of pRS315-SIR3 were constructed with gap repair using BmgBI/NdeI-linearized pRS315-SIR3. The two-hybrid constructs were generated by recombination of the mutant SIR3 fragments into EagI/NdeI-linearized pGAD-C2-SIR3 (307–978).
Purification of the Sir3 AAA+-like domain
A fragment corresponding to Sir3 amino acids 530–845 was subcloned into pET41. The construct was expressed in E. coli strain BL21 Rosetta pLysS and purified using the GST tag, and an integrated PreScission nuclease digestion sequence (Grum et al. 2010) was used to elute the recombinant Sir3 protein from the glutathione Sepharose. The resulting Sir3 (530–845) protein was further purified by Superdex 200 gel filtration and ion exchange chromatography. Protein concentrations were estimated by UV spectroscopy.
Crystallization and structure determination
The purified Sir3 AAA+ ATPase-like domain (amino acids 530–845) was concentrated to 9 mg/mL. Crystals grown at 4°C by hanging-drop vapor diffusion from mixtures containing equal volumes of protein and reservoir solutions containing 2 M ammonium sulfate, 2% (w/v) PEG-400, and 0.1 M HEPES (pH 7.5) were flash-frozen in mother liquor made up to 20% (v/v) glycerol. Diffraction data of SeMet crystals were collected on ID14-4 at ESRF and processed using XDS (Kabsch 2010) and SCALA (Evans 2006). SAD phases were calculated in SOLVE/RESOLVE (Terwilliger and Berendzen 1999), and the model was refined with REFMAC5 (Murshudov et al. 1997) and Phenix-Refine (Afonine et al. 2010) without NCS restraints throughout. Model building used COOT (Emsley and Cowtan 2004), and images were generated using PyMol (DeLano Scientific). Crystallographic statistics are in Supplemental Table II. Coordinates have been deposited in the Protein Data Bank database under accession code 3TE6.
Chromatin reconstitution, methylation, and binding assays
In vitro reconstitution of chromatin was carried out essentially as described (Cubizolles et al. 2006; Martino et al. 2009). Briefly, recombinant Xenopus laevis histones were used to reconstitute histone octamers, and chromatin was assembled in vitro by adding increasing amounts of purified histone octamer to a constant amount of Cy3-labeled 147-base-pair (bp) nonposition DNA, Cy5-labeled 147-bp 601-Widom sequence, or DNA arrays containing six 601-Widom positioning elements separated by 20 bp of linker DNA (Lowary and Widom 1998). Methylation of H3K79 was carried out on reconstituted chromatin using recombinant yeast Dot1 as described (Martino et al. 2009). Mass spectrometric analysis showed that H3K79 was mono-, di-, and trimethylated on 50%–70% of the available K79 residues (Frederiks et al. 2008; Martino et al. 2009). Full-length Sir3 was expressed by baculoviral infection of sf21 insect cells and was affinity-purified using a 6xHis tag (Cubizolles et al. 2006). The Sir3 AAA+ ATPase-like domain (AAA; amino acids 530–845) or an N-terminal truncation (AAAΔN; amino acids 545–845) were purified from E. coli as described above. Increasing amounts of the indicated Sir3 proteins were added to the nucleosomes, and after 10 min of incubation on ice, the samples were routinely run at 80 V for 90 min at 4°C on a 0.7% agarose gel. The Cy3- or Cy5-labeled DNA was visualized using a Typhoon 9400 scanner.
Baculovirus expression of Sir3-1067
A sir3-1067 EcoRI fragment was introduced into pVL1392-SIR3 (Ghidelli et al. 2001). Sf21 insect cells were infected with viruses for Sir2, Sir4, and Sir3 or Sir3-1067 as described (Cubizolles et al. 2006). To test for in vitro SIR complex assembly, Sir2/4 extracts were mixed with Sir3 or Sir3-1067 extracts and incubated for 3 h on calmodulin Sepharose beads (GE Healthcare) to allow Sir4–CBP binding and the assembly of Sir2–Sir3–Sir4 holocomplexes. Sir4–CBP was eluted, and the presence of coprecipitating Sir3 was tested by Western blotting against the Sir3-HA tag. Sir4 and Sir2 were monitored with an anti-CBP antibody and an anti-Sir2 antibody (Santa Cruz Biotechnology), respectively.
Acknowledgments
We thank R. Kamakaka, D. Shore, L. Pillus, and F. van Leeuwen for strains and reagents; R. Schwegmann, L. Neumann, and K. Ruf for help with allele constructions; C. Vole, A. Rüppel, J. Wohlgemuth, M. Rübeling, and K. Nicklasch for technical assistance; V. Rybin for help with ITC; the staff at ESRF Grenoble for technical support at the synchrotron; and all members of our laboratories for discussions. This work was supported by the University of Duisburg-Essen (to A.E.M.), the European Molecular Biology Laboratory (to A.G.L.), the Human Frontiers Science Program (to A.G.L.), and the Deutsche Forschungsgemeinschaft (DFG grant EH237/6-1 to A.E.M. and LA 2489/1-1 to A.G.L.). S.K. was supported by an FWF-Schroedinger Fellowship. The Gasser laboratory is supported by the Novartis Research Foundation and the Marie Curie Training network, 4D Nucleosome.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.17175111.
References
- Afonine PV, Mustyakimov M, Grosse-Kunstleve RW, Moriarty NW, Langan P, Adams PD 2010. Joint X-ray and neutron refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 66: 1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altaf M, Utley RT, Lacoste N, Tan S, Briggs SD, Cote J 2007. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol Cell 28: 1002–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Mitchell J, Leber J, Kobayashi R, Stillman B 1995. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell 83: 563–568 [DOI] [PubMed] [Google Scholar]
- Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev 7: 592–604 [DOI] [PubMed] [Google Scholar]
- Buchberger JR, Onishi M, Li G, Seebacher J, Rudner AD, Gygi SP, Moazed D 2008. Sir3–nucleosome interactions in spreading of silent chromatin in Saccharomyces cerevisiae. Mol Cell Biol 28: 6903–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Rai R, Zhou ZR, Kanoh J, Ribeyre C, Yang Y, Zheng H, Damay P, Wang F, Tsujii H, et al. 2011. A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat Struct Mol Biol 18: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CC, Li YC, Gartenberg MR 2008. Bypassing Sir2 and O-acetyl-ADP-ribose in transcriptional silencing. Mol Cell 31: 650–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M, Palladino F, Laroche T, Kyrion G, Liu C, Lustig AJ, Gasser SM 1995. The carboxy termini of Sir4 and Rap1 affect Sir3 localization: evidence for a multicomponent complex required for yeast telomeric silencing. J Cell Biol 129: 909–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JJ, Yuan P, Hsu HC, Li Z, Xu RM, Sternglanz R 2006. Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain. Mol Cell Biol 26: 3256–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubizolles F, Martino F, Perrod S, Gasser SM 2006. A homotrimer–heterotrimer switch in Sir2 structure differentiates rDNA and telomeric silencing. Mol Cell 21: 825–836 [DOI] [PubMed] [Google Scholar]
- Dueber EL, Corn JE, Bell SD, Berger JM 2007. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science 317: 1210–1213 [DOI] [PubMed] [Google Scholar]
- Ehrenhofer-Murray AE 2004. Chromatin dynamics at DNA replication, transcription and repair. Eur J Biochem 271: 2335–2349 [DOI] [PubMed] [Google Scholar]
- Ehrentraut S, Weber JM, Dybowski JN, Hoffmann D, Ehrenhofer-Murray AE 2010. Rpd3-dependent boundary formation at telomeres by removal of Sir2 substrate. Proc Natl Acad Sci 107: 5522–5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Evans P 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62: 72–82 [DOI] [PubMed] [Google Scholar]
- Frederiks F, Tzouros M, Oudgenoeg G, van Welsem T, Fornerod M, Krijgsveld J, van Leeuwen F 2008. Nonprocessive methylation by Dot1 leads to functional redundancy of histone H3K79 methylation states. Nat Struct Mol Biol 15: 550–557 [DOI] [PubMed] [Google Scholar]
- Gasser SM, Cockell MM 2001. The molecular biology of the SIR proteins. Gene 279: 1–16 [DOI] [PubMed] [Google Scholar]
- Georgel PT, Palacios DeBeer MA, Pietz G, Fox CA, Hansen JC 2001. Sir3-dependent assembly of supramolecular chromatin structures in vitro. Proc Natl Acad Sci 98: 8584–8589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghidelli S, Donze D, Dhillon N, Kamakaka RT 2001. Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities. EMBO J 20: 4522–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762 [DOI] [PubMed] [Google Scholar]
- Grum D, van den Boom J, Neumann D, Matena A, Link NM, Mueller JW 2010. A heterodimer of human 3′-phospho-adenosine-5′-phosphosulphate (PAPS) synthases is a new sulphate activating complex. Biochem Biophys Res Commun 395: 420–425 [DOI] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80: 583–592 [DOI] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M 1996. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383: 92–96 [DOI] [PubMed] [Google Scholar]
- Hickman MA, Rusche LN 2010. Transcriptional silencing functions of the yeast protein Orc1/Sir3 subfunctionalized after gene duplication. Proc Natl Acad Sci 107: 19384–19389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe GJ, Tanny JC, Rudner AD, Gerber SA, Danaie S, Gygi SP, Moazed D 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol 22: 4167–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Danzer JR, Fox CA, Keck JL 2006. Structure of the Sir3 protein bromo adjacent homology (BAH) domain from S. cerevisiae at 1.95 Å resolution. Protein Sci 15: 1182–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800 [DOI] [PubMed] [Google Scholar]
- Johnson LM, Kayne PS, Kahn ES, Grunstein M 1990. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc Natl Acad Sci 87: 6286–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W 2010. Xds. Acta Crystallogr D Biol Crystallogr 66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DA, Hall BE, Iwamoto MA, Win KZ, Chang JF, Ellenberger T 2006. Domain structure and protein interactions of the silent information regulator Sir3 revealed by screening a nested deletion library of protein fragments. J Biol Chem 281: 20107–20119 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Liaw H, Lustig AJ 2006. Sir3 C-terminal domain involvement in the initiation and spreading of heterochromatin. Mol Cell Biol 26: 7616–7631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D 2005. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 121: 515–527 [DOI] [PubMed] [Google Scholar]
- Lowary PT, Widom J 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol 276: 19–42 [DOI] [PubMed] [Google Scholar]
- Luo K, Vega-Palas MA, Grunstein M 2002. Rap1–Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev 16: 1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino F, Kueng S, Robinson P, Tsai-Pflugfelder M, van Leeuwen F, Ziegler M, Cubizolles F, Cockell MM, Rhodes D, Gasser SM 2009. Reconstitution of yeast silent chromatin: multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol Cell 33: 323–334 [DOI] [PubMed] [Google Scholar]
- McBryant SJ, Krause C, Hansen JC 2006. Domain organization and quaternary structure of the Saccharomyces cerevisiae silent information regulator 3 protein, Sir3p. Biochemistry 45: 15941–15948 [DOI] [PubMed] [Google Scholar]
- Megee PC, Morgan BA, Mittman BA, Smith MM 1990. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science 247: 841–845 [DOI] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev 8: 2257–2269 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV 1999. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res 9: 27–43 [PubMed] [Google Scholar]
- Norris A, Boeke JD 2010. Silent information regulator 3: the Goldilocks of the silencing complex. Genes Dev 24: 115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris A, Bianchet MA, Boeke JD 2008. Compensatory interactions between Sir3p and the nucleosomal LRS surface imply their direct interaction. PLoS Genet 4: e1000301 doi: 10.1371/journal.pgen.1000301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D 2007. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell 28: 1015–1028 [DOI] [PubMed] [Google Scholar]
- Oppikofer M, Kueng S, Martino F, Soeroes S, Hancock SM, Chin JW, Fischle W, Gasser SM 2011. A dual role of H4K16 acetylation in the establishment of yeast silent chromatin. EMBO J 10: 2610–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EC, Szostak JW 1990. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol Cell Biol 10: 4932–4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J, Herskowitz I 1987. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics 116: 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner AD, Hall BE, Ellenberger T, Moazed D 2005. A nonhistone protein–protein interaction required for assembly of the SIR complex and silent chromatin. Mol Cell Biol 25: 4514–4528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol Biol Cell 13: 2207–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem 72: 481–516 [DOI] [PubMed] [Google Scholar]
- Sampath V, Yuan P, Wang IX, Prugar E, van Leeuwen F, Sternglanz R 2009. Mutational analysis of the Sir3 BAH domain reveals multiple points of interaction with nucleosomes. Mol Cell Biol 29: 2532–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F 1991. Getting started with yeast. Methods Enzymol 194: 3–21 [DOI] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, et al. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci 97: 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EM, Reifsnyder C, McVey M, Gazo B, Pillus L 2000. Two classes of sir3 mutants enhance the sir1 mutant mating defect and abolish telomeric silencing in Saccharomyces cerevisiae. Genetics 155: 509–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner KG, Landry J, Sternglanz R, Denu JM 2000. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci 97: 14178–14182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J 1999. Evaluation of macromolecular electron-density map quality using the correlation of local r.m.s. density. Acta Crystallogr D Biol Crystallogr 55: 1872–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JS, Snow ML, Giles S, McPherson LE, Grunstein M 2003. Identification of a functional domain within the essential core of histone H3 that is required for telomeric and HM silencing in Saccharomyces cerevisiae. Genetics 163: 447–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745–756 [DOI] [PubMed] [Google Scholar]
- Woodcock CL 2006. Chromatin architecture. Curr Opin Struct Biol 16: 213–220 [DOI] [PubMed] [Google Scholar]
- Zachariae W, Shevchenko A, Andrews PD, Ciosk R, Galova M, Stark MJ, Mann M, Nasmyth K 1998. Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to cullins. Science 279: 1216–1219 [DOI] [PubMed] [Google Scholar]