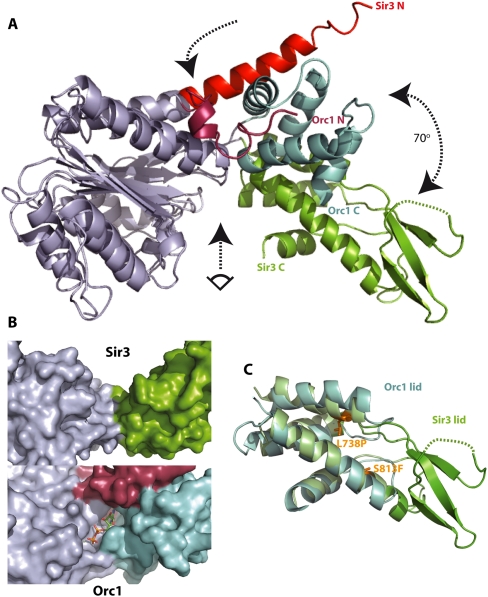
Figure 2.
The Sir3 AAA+ ATPase-like domain has a strikingly different conformation from its closest structural relative, Orc1/cdc6, and does not contain a nucleotide-binding pocket. (A) Superposition of the AAA+-like domains of Sir3 and of S. solfataricus ORC1/cdc6 (2qby, A). Structural alignments were produced by superposition of the residues within the base subdomain of the two structures (RMSD of Cα atoms < 1.6 Å). For clarity, base subdomains are both colored gray. Other subdomain features are colored differently to highlight differences. The arrows indicate the hinge region (top arrow), the rotation between the lid domains of Sir3 and ORC1/Cdc6 (right arrow), and the view into the (potential) nucleotide-binding pocket shown in B (bottom arrow). (B) Surface representation of the presumed nucleotide-binding pocket of Sir3 (top) compared with the ADP bound nucleotide-binding pocket in S. solfataricus ORC1 (bottom). The shallow and wide groove in Sir3 is not compatible with a suggested OAADPR-binding function. (C) Superposition of the lid subdomains of Sir3 and of S. solfataricus ORC1 (2qby, A). Structural alignments were produced by superposition of the helical residues within the lid subdomains of the two structures (RMSD of Cα atoms = 2.1 Å). Additional structural features in the Sir3 lid subdomain are evident (green). These include a threefold anti-parallel β sheet containing mostly positively charged residues. The location of two known point mutants causing a sir3 phenotype (Stone et al. 2000; Buchberger et al. 2008) are also indicated.