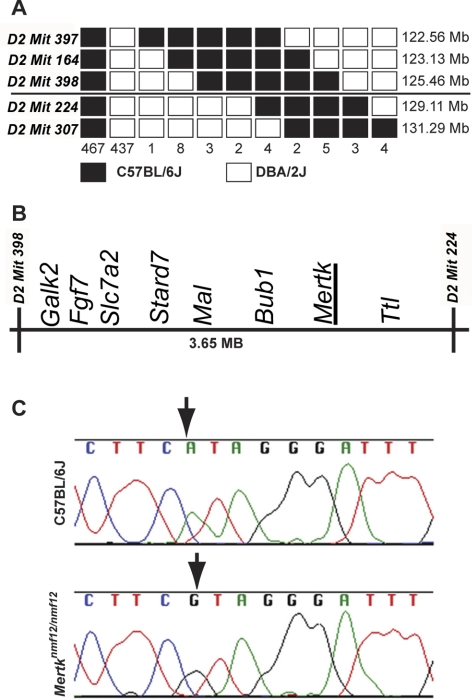
Figure 1.
Mapping and cloning of the nmf12 mutation. (A) A haplotype map is shown, representing the location and number of crossovers in 936 meioses observed in 468 affected and unaffected progeny-tested mice. The numbers at the bottom of the graph represent the number of chromosomes bearing each depicted crossover. The critical interval spans 3.65 Mb between markers D2Mit398 (125.46 Mb) and D2Mit224 (129.11 Mb). Within this critical region (B) lies Mertk, a gene in which mutations have been shown to cause retinal degeneration. (C) Sequence chromatograms from control, C57BL/6J, and Mertknmf12 homozygotes. Analysis of these sequences reveals an A-to-G transition at base pair 2237 (A2237G).