A new interaction is reported between lens aquaporin-0 and the ezrin/radixin/moesin family of proteins that form links with the actin cytoskeleton. Aquaporin-0 may be the missing link in specialized membrane protein–cytoskeleton protein complexes in the lens.
Abstract
Purpose.
Aquaporin 0 (AQP0) is the major intrinsic protein in the lens and is essential for establishing proper fiber cell structure and organization. Cytoskeletal proteins that directly interact with the C terminus of AQP0 are identified herein.
Methods.
The water-insoluble fraction of lens fiber cells was chemically cross-linked, and cross-linked peptides with the C terminus of AQP0 were identified by mass spectrometry. Coimmunoprecipitation and AQP0 C-terminal peptide pulldown experiments were used to confirm the protein–protein interaction.
Results.
Unexpectedly, AQP0 was found to directly associate with ezrin/radixin/moesin (ERM) family members, proteins that are involved in linkage of actin filaments to the plasma membrane. Cross-linked peptides were detected between AQP0 and degenerate sequences of ezrin and radixin; however, AQP0 interaction with ezrin is believed to play a more significant function in the lens because of its higher level of expression and observed ezrin-specific cross-linking. The interaction was found to occur between the C terminus of AQP0 and subdomains F1 and F3 of ERM proteins. The interaction between AQP0 and ezrin was confirmed by coimmunoprecipitation and AQP0 C-terminal peptide pulldown experiments.
Conclusions.
Considering the important known functions of the cellular actin cytoskeleton in fiber cell differentiation, the interaction of AQP0 and ERM proteins may play an important role in fiber cell morphology, elongation, and organization.
The lens of the eye is a transparent tissue composed of an anterior monolayer of epithelial cells and the underlying highly differentiated and elongated lens fiber cells that form the bulk of the organ. Lens transparency is critical to its function of focusing of light onto the retina, and this transparency is achieved and maintained by precise cell–cell interactions. Lens fiber cells are closely packed together with extensive contact between adjacent cells forming cell-to-cell adhesive complexes, the cortex adhaerens.1 Disruption of fiber cell packing can lead to light scattering and cataract.2
Aquaporin 0 (AQP0), the major intrinsic membrane protein of the lens, is the most abundant membrane protein in the lens, constituting 50% to 60% of the plasma membrane protein,3 and it plays important roles in the maintenance of lens transparency and homeostasis. Mutation in and knockout of the AQP0 gene results in lens cataract.4–7 AQP0 has been suggested to perform multiple roles in the lens, including functioning as a water channel4,8,9 and as a cell adhesion molecule.10–15
AQP0 has six transmembrane domains, with its N terminus and C terminus located in the cytoplasm. The cytoplasmic C terminus of AQP0 is predicted to be functionally significant because it contains sites of modifications and protein–protein interactions. The C terminus of AQP0 contains several phosphorylation sites16,17 and a calmodulin interaction site,18 all of which have been reported to impact AQP0 permeability.19,20 The C terminus of AQP0 has also been reported to interact with the cytoskeletal proteins filensin and CP49, suggesting a role for AQP0 in establishing and/or maintaining fiber cell shape.21
The ezrin/radixin/moesin (ERM) protein family is part of the band 4.1 super family, which links actin filaments to the plasma membrane.22–24 Specific ERM proteins are involved in many important roles, such as cell signaling, stabilizing adhesion junctions, the maintenance of cell shape, villar organization in the gut, and the light-regulated maintenance of photoreceptors, etc.25 ERM protein members all share a common homologous ∼300–amino acid domain called the FERM domain (F for 4.1 protein, E for ezrin, R for radixin, and M for moesin).26 ERM proteins bind with membrane proteins such as, CD44, intracellular adhesion molecule-1 and −3, P-selectin glycoprotein ligand-1, and others through an N-terminal FERM domain and connect to F-actin through a C-terminal domain.
The presence of ERM proteins in lens fiber cells has been shown1,27–29; however, the function of ERM proteins in lens fiber cells has not been extensively studied. Ezrin was identified as one of the components of a novel cell–cell junction system of lens fiber cells, which contains ezrin, periplakin, periaxin, and desmoyokin (EPPD complex).1 It is unclear how the EPPD complex is linked to the plasma membrane, because the transmembrane protein in this complex has not been determined. In this article, we report a direct interaction between the FERM domain of ezrin and AQP0. The results indicate that AQP0 is a candidate membrane protein attachment site of the EPPD complex.
Materials and Methods
Preparation of Water Insoluble Fraction
Frozen 1-year-old or older bovine lenses (PelFreez Biologicals, Rogers, AK) were decapsulated and dissected into cortex and nucleus before homogenization. Tissue was homogenized in homogenizing buffer (25 mM Tris buffer containing 5 mM EDTA, 1 mM PMSF, and 150 mM NaCl, pH 8.0) and centrifuged at 88,000 g for 20 minutes, and the supernatant was discarded. The pellets were washed three times with homogenizing buffer. The remaining pellets are called the water insoluble fraction (WIF).
Cross-Linking Reactions
The WIF was washed twice with 10 mM sodium phosphate buffer containing 100 mM NaCl, pH 7.4. The remaining pellets were resuspended in 1 mL of phosphate buffer and 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) was added to the solution to a final concentration of 5 mM. The mixture was incubated at room temperature for 1 hour. Excess EDC was removed by centrifuging at 88,000 g, and the remaining pellets were washed three times with water followed by three washes with 8 M urea and one wash with 0.1 M sodium hydroxide. The remaining pellets were further washed with water and called the cross-linked membrane fraction.
Coimmunoprecipitations
The WIF was suspended in Tris buffer (25 mM Tris, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 1mM PMSF, and 0.1% SDS, pH 7.4) and centrifuged for 15 minutes at 80,000 g at 4°C. A mouse ezrin antibody (10 μL of 3C12; Sigma Chemical, Saint Louis, MO) was then added to the supernatant and incubated at 4°C overnight followed by incubation with protein G–coated beads at room temperature for 2 hours. A control sample was prepared by incubating the supernatant with protein G beads alone without antibody or protein G beads with control mouse immunoglobulin G. The beads were washed ten times with the above buffer and bound proteins were eluted with 40 μL of SDS sample buffer (Invitrogen, Carlsbad, CA). Samples were loaded onto a 4–12% NuPAGE Novex Bis-Tris gel and MOPS running buffer was used for separation (Invitrogen). For each sample, two lanes were used, and 20 μL was loaded on each lane. After SDS-PAGE, the gel was cut in half, and half of the gel was Coomassie-stained and the visible gel bands were excised for in-gel trypsin digestion. The other half of the gel was used for immunoblotting. Gel-separated proteins were electrophoretically transferred from the gel to a nitrocellulose membrane. The membrane was blocked with 5% nonfat milk in Tris-buffered saline and Tween 20 (TBST) for 1 hour. Blots were then incubated with the rabbit anti-AQP0 primary antibody in TBST overnight at 4°C followed by six TBST washes for 15 minutes each. Goat anti-rabbit secondary antibody (1 μL) labeled with Dylight 680 (Pierce, Rockford, IL) was added and incubated for 1 hour at room temperature followed by six TBST washes for 15 minutes each. The results were read using an Odyssey Infrared Imager (LI-COR Biosciences, Lincoln, NE). Similarly, the membrane was further blotted with mouse anti-ezrin primary antibody and goat anti-mouse Dylight 800 (Pierce). To identify other proteins in the immunoprecipitation samples, the eluate from the beads conjugated with mouse immunoglobulin G or beads conjugated with ezrin antibody was run into the gel for 1.5 to 2 cm. Then the gel was Coomassie-stained and the whole stained area was excised and diced to 1 mm cubes for in-gel digestion.
Trypsin Digestion
The cross-linked membrane fraction was reduced in 10 mM DTT at 56°C for 1 hour and alkylated in 55 mM iodoacetamide at room temperature in the dark for 45 minutes. The reduced and alkylated membrane fraction was washed with water twice and resuspended in 50 mM ammonium bicarbonate buffer (pH 8.0). Trypsin (1 μg) was used to digest the cross-linked membrane fraction from one bovine lens cortex. The digestion was performed at 37°C for 18 hours. The cross-linked membrane fraction was further digested for another 18 hours at 37°C by adding 0.5 μg of trypsin to achieve a more complete digestion. After digestion, the solution was dried in a SpeedVac (Thermo Fisher Scientific, Waltham, MA).
Immunoprecipitated samples were separated by SDS-PAGE as described above and gel bands were excised for in-gel digestion. For in-gel digestion, the gel bands were destained with three consecutive washes with a 50:50 mixture of 50 mM ammonium bicarbonate and acetonitrile for 10 minutes. The gel bands were dried in a SpeedVac. Each sample containing an individual band was rehydrated in a 10- to 15-μL solution containing 20 ng/μL trypsin (Promega, Madison, WI) in 50 mM ammonium bicarbonate for 15 minutes; 30 μL of 50 mM ammonium bicarbonate buffer was added to each sample and the samples were incubated at 37°C for 18 hours. Peptides were extracted using 20%ACN/0.1%TFA once, 60%ACN/0.1%TFA twice, and 80%ACN/0.1%TFA once. The extracted samples were pooled and dried in a SpeedVac and reconstituted in 0.1% formic acid for subsequent liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis.
Fractionation of Tryptic Peptides by Offline Strong Cation Exchange
Tryptic peptides of the cross-linked membrane fraction were resuspended in 5 mM potassium phosphate buffer containing 30% ACN, pH 2.5 (buffer A). The solution was centrifuged at 20,000 g for 15 minutes and the supernatant was collected and added to strong cation exchange resin (Luna SCX, 5 μm, 100 Å media) in a spin cup. After 15 minutes of incubation, unbound peptides were collected by centrifugation at 1000 g. The resin was washed by buffer A twice and bound peptides were step-eluted sequentially by 40%, 60%, and 100% buffer B (5 mM potassium phosphate buffer containing 30% ACN, 350 mM KCl, pH 2.5) balanced with buffer A. The 60% and 100% buffer B eluate were dried in a SpeedVac and reconstituted in 0.1% formic acid. The peptides were desalted using a C18 Ziptip (Millipore, Billerica, MA) and eluted in 70% ACN (0.1% formic acid). The eluate was dried in a SpeedVac and reconstituted in 0.1% formic acid for further analysis.
LC/Electrospray Ionization MS/MS
Tryptic peptides were either directly separated on a 1-dimensional fused silica capillary column (150 mm × 100 μm) packed with Phenomenex Jupiter resin (3 μm mean particle size, 300 Å pore size) or analyzed by multidimensional protein identification technology (MudPIT).30 For cross-linked peptide identification, the 60% and 100% buffer B eluates from off-line strong cation exchange fractionation were analyzed on an LTQ Orbitrap mass spectrometer (ThermoFisher, San Jose, CA). Cross-linked peptides were identified in both fractions; however, interpretable tandem mass spectra were not always obtained from potential cross-linked peptides because of low signal-to-noise ratios. Therefore, the 60% and 100% buffer B eluates were combined and were subjected to MudPIT analysis as described below.
One-dimensional LC was performed on both cross-linked samples and in-gel digested immunoprecipitated samples using the following gradient at a flow rate of 0.5 μL per minute: 0 to 10 minutes, 2% ACN (0.1% formic acid); 10 to 50 minutes, 2% to 35% ACN (0.1% formic acid); and 50 to 60 minutes, 35% to 90% ACN (0.1% formic acid) balanced with 0.1% formic acid. The eluate was directly infused into an LTQ Orbitrap (for cross-linked samples) or an LTQ Velos mass spectrometer for immunoprecipitated samples (ThermoFisher, San Jose, CA) equipped with a nanoelectrospray source. For MudPIT analysis, peptides were pressure-loaded onto a custom packed biphasic C18/SCX trap column (6 cm × 150 μm, Jupiter C18, 5 μm, 300 Å media followed by 8 cm × 150 μm, Luna SCX, 5 μm, 100 Å media). The trap column was coupled to a nanoflow analytical column (11 cm × 100 μm, Jupiter C18, 3 μm, 300 Å media). MudPIT analysis was performed with a nine-step salt pulse gradient (100 mM, 200 mM, 500 mM, 750 mM, 1 M, 1.5 M, 2 M, and 3 M ammonium acetate). Peptides were eluted from the analytical column after each salt pulse with a 90-minute reversed-phase solvent gradient (2% to 45% ACN containing 0.1% formic acid) for the first seven salt pulses and a 90-minute reversed-phase solvent gradient (2% to 95% ACN containing 0.1% formic acid) for the last two salt pulses. The eluate was directly infused into an LTQ Velos mass spectrometer. The instruments were operated in a data-dependent mode for all the analyses with the top five most abundant ions in each MS scan selected for fragmentation in the LTQ.
Data Analysis
For identifying the cross-linked peptides, the raw data were manually interpreted or converted to Mascot generic format (MGF) files by an in-house developed algorithm, Scansifter, and cross-linked peptides were identified with the help of the xQuest software (IMSB, Zurich Institute of Molecular Systems Biology).31 MS data were searched with mass tolerance of 5 ppm (for LTQ-Orbitrap) and for MS/MS data a maximum mass error of 0.5 u was allowed. For database searching, all MS/MS spectra were converted to data files by Scansifter and searched against a concatenated forward and reversed bovine Swissprot database (August 2010). Results were filtered to a 5% peptide false discovery rate with the requirement for a minimum of two peptides per reported protein using IDPicker.32
AQP0 C-Terminal Peptide Affinity Pulldown
A bovine AQP0 peptide 240 to 259 (CSRPSESDGQPEVTGEPVELK) was synthesized by EZBiolab (Carmel, IN). Cysteine was added to the N terminus of the peptide for easy immobilization on cysteine containing beads and residue 246 is changed to its deamidated form of Asp because a majority of AQP0 is found deamidated at residue 246. Homologous sequences of the AQP0 loop region containing residues 110 to 127 (CPAVRGNLALNTLHPAVSV) and the N terminus residues 1 to 9 (MWELRSASFC) were synthesized at the MUSC proteogenomics facility. Half of a milligram of each peptide was solubilized in 200 μL 50 mM Tris buffer, 5 mM EDTA, pH 8.0. The solution was added to 500 μL of immobilized Tris(2-carboxyethyl)phosphine (TCEP) in a Handee Spin Cup column (Pierce, Rockford, IL) to reduce any disulfide bonds formed and incubated at room temperature for 1 hour. The peptide solution was collected by centrifugation, and the immobilized TCEP beads were washed by 300 μL Tris buffer and the wash solution was combined with the peptide solution. 0.2 mg EZ-link idoacetyl-PEG2-biotin (Pierce, Rockford, IL) was added to the peptide solution immediately to label the peptide with biotin. The reaction mixture was incubated at room temperature for 1 hour in the dark. Biotinylation of the peptides was confirmed by MALDI mass spectrometry (Bruker Autoflex III; Bruker Daltonik, Bremen, Germany) and the level of reaction was controlled to reach at least 50%, but < 90% to avoid excess biotinylation regent in the reaction mixture. The reaction was allowed to process for 1 more hour at room temperature in the dark and then added to 100 μL of streptavidin beads (Pierce, Rockford, IL). The beads were incubated at room temperature for 30 minutes and washed 10 times by 25 mM Tris, 5 mM EDTA, 1 mM PMSF, 10 mM NaCl, and 1 mM MgCl2. The WIF prepared as described above was extracted by 25 mM Tris, 5 mM EDTA, 1 mM PMSF, 150 mM NaCl, and 1 M KCl. The salt in the extract was removed by filtration through 10-kDa molecular weight cutoff filter (Millpore, Billerica, MA). The WIF extract was solubilized in 25 mM Tris, 5 mM EDTA, 1 mM PMSF, 10 mM NaCl, and 1 mM MgCl2 and loaded onto AQP0 peptide-conjugated streptavidin beads (200 μL). The beads were incubated at 4°C overnight followed by washes (6×) with 25 mM Tris, 5 mM EDTA, 1 mM PMSF, 10 mM NaCl, and 1 mM MgCl2 and washes (2×) with 25 mM Tris, 5 mM EDTA, 1 mM PMSF, 10 mM NaCl, 1 mM MgCl2, and 1% triton. The bound proteins on the beads were eluted by 30 μL of SDS sample buffer and subjected to SDS-PAGE and Western blotting for ezrin using mouse anti-ezrin primary antibody (3C12; Sigma, Saint Louis, MO) and goat anti-mouse Dylight 800 (Pierce).
Results
Cross-Linking Reaction and Enrichment of Cross-Linked Peptides
EDC (1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride) is a zero-length cross-linking reagent used to couple carboxyl groups to primary amines. The cross-linking reaction results in a stable amide bond and release of H2O. The EDC cross-linked membrane fraction contains a complicated mixture of large protein complexes. Moreover, the cross-linking reaction normally occurs at a substoichiometric level. Identification of cross-linked peptide from this complicated mixture is challenging; therefore, cross-linked peptide enrichment was performed before MS analysis. Cross-linked tryptic peptides have at least four basic groups (two N-terminal amino groups and two C-terminal Lys/Arg residues); therefore, cross-linked peptides bind stronger on strong cation exchange resin than regular tryptic peptides. In this study, we enriched the cross-linked peptides via offline strong cation exchange.
The 60% buffer B eluate from offline strong cation exchange was analyzed with a LTQ Orbitrap mass spectrometer. Cross-linked peptides containing the C terminus of AQP0 were searched by xQuest. In addition, MS/MS analysis of the bovine AQP0 C-terminal peptide 239 to 259 produces intense fragment ion signals (m/z: 585, 771, 872, and 971); therefore, tandem mass spectra containing these fragment ions were also thoroughly searched manually. For cross-linked peptide identification, peptides with +1 or +2 charges were ignored. In addition, tryptic cleavage C-terminal to cross-linked lysine residues was not considered. Only two proteins were found to be cross-linked to the AQP0 C terminus: (1) the previously reported AQP0 interaction partner filensin21 and (2) the ERM proteins ezrin and radixin. No other proteins were detected to cross-link with the C terminus of AQP0. Only the cross-linked peptides between ezrin and AQP0 are presented in this article.
Identification of Cross-Linked Peptides between ERM Proteins and the C Terminus of AQP0
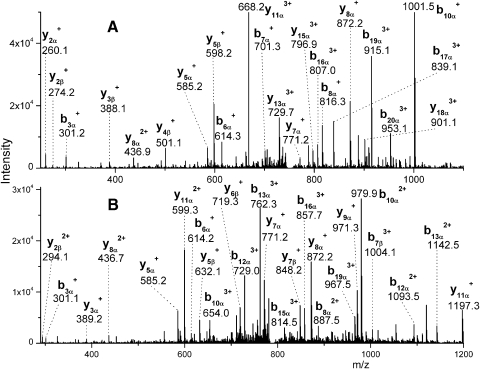
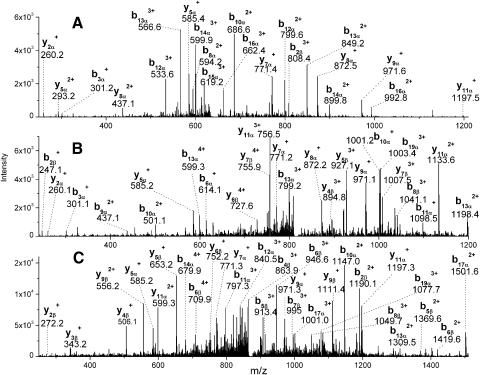
Five cross-linked peptides between AQP0 and ERM sequences were repeatedly identified in multiple experiments based on highly accurate parent mass measurements and their tandem mass spectra in the cross-linked membrane fraction from lens cortex. These cross-linked peptides were not detected in the cross-linked membrane fraction from the lens nucleus. Table 1 lists these five identified peptides. The measured masses of these peptides are within 5 ppm of the predicted masses. Parent ions of these peptides were detected by 1D LC-MS/MS analysis, and the first three cross-linked peptides were also confirmed by their tandem mass spectra. The tandem mass spectra of the last two cross-linked peptides were weak by 1D LC-MS/MS. Therefore, the sample was analyzed by a more thorough MudPIT analysis to confirm the sequences, and their tandem mass spectra were obtained. Five tandem mass spectra corresponding to five different ERM peptides are shown in Figure 1 and Figure 2. Note that characteristic, strong fragment ions (m/z: 585, 771, 872, and 971) of the AQP0 C-terminal tryptic peptide (239–259) are observed in each MS/MS spectrum and this pattern facilitated the identification of AQP0 cross-linked peptides. With the exception of the cross-linked peptide between AQP0 239 to 259 with the short ERM peptide NKK, the tandem mass spectra of other cross-linked peptides have a continuous series of b- or y- ions from ERM peptides and strong y- ions corresponding to cleavage of proline amide bonds.
Table 1.
Identified Cross-Linked Peptides between Ezrin and AQP0 C-Terminus
| Predicted (MH)+ | Observed (MH)+ | Charge | Error (ppm) | Cross-Linked Peptide |
|---|---|---|---|---|
| 3002.554 | 3002.548 | 4 | −1.9 | AQP0: GSRPSESDGQPE*VTGEPVELK |
| 3002.544 | 5 | −3.5 | ERM: PK*PINVR | |
| 3155.585 | 3155.581 | 4 | −3.2 | AQP0: GSRPSE*SD*GQPEVTGEPVELK |
| 3155.579 | 5 | −2.8 | ERM: K*ESPLQFK | |
| 2568.290 | 2568.281 | 4 | −1.4 | AQP0: GSRPSE*SD*GQPEVTGEPVELK |
| 2568.282 | 3 | −1.9 | ERM: NK*K | |
| 3266.727 | 3266.718 | 4 | −2.8 | AQP0: GSRPSESD*GQPE*VTGEPVELK |
| 3266.719 | 5 | −2.4 | ERM: FVIKPIDK*K | |
| 3266.719 | 6 | −2.4 | ||
| 3489.728 | 3489.724 | 4 | −1.0 | AQP0: GSRPSE*SD*GQPEVTGEPVELK |
| 3489.719 | 5 | −2.6 | ERM: K*APDFVFYAPR |
Listed masses are monoisotopic masses.
Residues that are involved in crosslinking. If two asterisks are on one peptide, crosslinking residues cannot be unambiguously assigned or crosslinking can occur on either residue.
Figure 1.
Tandem mass spectra of two cross-linked peptides between AQP0 and ezrin subdomain F1. (A) Spectrum of quadruply charged ion corresponding to AQP0 239 to 259 (GSRPSESDGQPEVTGEPVELK) cross-linked with ERM residues 2 to 8 (PKPINVR) (m/z 751.4). (B) Spectrum of quadruply charged ion corresponding to AQP0 239 to 259 cross-linked with ezrin 72 to 79 (KESPLQFK) (m/z 790.3). Annotation of the cross-linked peptides as α- and β-chains is performed according to Schilling et al.,33 with AQP0 239 to 259 assigned to the α-chain and ERM peptides assigned to the β-chain. Only the b- and y-ions are labeled.
Figure 2.
Tandem mass spectra of three cross-linked peptides between AQP0 and ERM subdomain F3. (A) Spectrum of quadruply charged ion corresponding to AQP0 239 to 259 cross-linked with ERM residues 210 to 212 (NKK) (m/z 643.3). (B) Spectrum of quadruply charged ion corresponding to AQP0 239 to 259 cross-linked with ERM residues 255 to 263 (FVIKPIDKK) (m/z 818.0). (C) Spectrum of quadruply charged ion corresponding to AQP0 239 to 259 cross-linked with ERM residues 263 to 273 (KAPDFVFYAPR) (m/z 873.0). Annotation of the cross-linked peptides as α- and β-chains is performed according to Schilling et al.,33 with AQP0 239 to 259 assigned to the α-chain and and ezrin peptide assigned to the β-chain. Only the b- and y-ions were labeled.
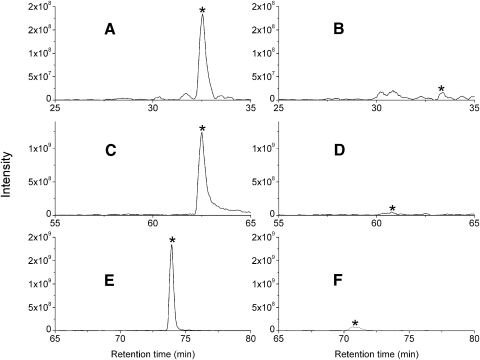
The ERM proteins are highly similar paralogs (∼75% sequence identity).34 Both ezrin and radixin are present in lens fiber cells.28,29 Moesin was not detected by Bagchi et al.28 in lens fiber cells, but Rao et al.29 reported the presence of moesin in lens fiber cells. To confirm the presence of three members of ERM proteins, the water insoluble fraction was extracted by 8 M urea and the urea soluble fraction was separated by SDS-PAGE as previously reported.35 The 80-kDa gel band was digested by trypsin and the tryptic peptides were analyzed by LC-MS/MS. Ezrin- and radixin-specific peptides, but no moesin-specific peptides were detected by MS, suggesting that moesin is not present or at least is in very low abundance in the bovine lens. Except for the peptide KESPLQFK of ezrin, the other four peptides involved in cross-linking with AQP0 are present in both ezrin and radixin. The ezrin peptide KESPLQFK has the sequence of KENPLQFK in radixin. A weak tandem mass spectrum of cross-linked peptide between AQP0 239 to 259 and radixin 72 to 79 (KENPLQFK) was also detected which indicates that AQP0 also interacts with radixin (data not shown). The relative abundance of ezrin and radixin peptides in the 80-kDa gel band digest was analyzed. The selected ion chromatograms of three paired peptides from ezrin and radixin are shown in Figure 3. Each peptide pair differs by only a few residues; therefore, their ionization efficiency is expected to be similar. These data indicate that ezrin is much more abundant than radixin in lens fiber cells.
Figure 3.
The selected ion chromatograms of the paired peptides of ezrin and radixin from LC-MS/MS analysis of the 80-kDa band in the urea soluble fraction. (A) Ezrin 72 to 79 (KESPLQFK); (B) radixin 72 to 79 (KENPLQFK); (C) ezrin 194 to 209 (IAQDLEMYGINYFEIK); (D) radixin 194 to 209 (IAQDLEMYGVNYFEIK); (E) ezrin 84 to 100 (FYPEDVAEELIQDITQK); and (F) radixin 84 to 100 (FFPEDVSEELIQEITQR). Asterisks indicate the peaks of interest.
The observed cross-linked peptides contain the C terminus of AQP0 and peptides from the ERM FERM domain. Cross-linking between other regions of AQP0 and the ERM proteins was not detected. The ERM FERM domain is a clover-shaped structure consisting of three structural domains (subdomain F1, F2, and F3).37 Figure 4 shows the crystal structure of the ezrin FERM domain (PDB entry 1NI2) with cross-linked residues labeled. The cross-linked ezrin peptides 2 to 8 (PKPINVR) and 72 to 79 (KESPLQFK) are located in subdomain F1, and peptides 210 to 212 (NKK), 255 to 263 (FVIKPIDKK) and 263 to 273 (KAPDFVFYAPR) are located in subdomain F3. Based on the crystal structure of the FERM domain, lysine residues K3 and K72 in subdomain F1 are in close proximity to each other, as are lysine residues K211, K262, and K263 in subdomain F3;36 however, subdomains F1 and F3 are not close enough to interact with the same region of AQP0. One possible explanation is that separate ERM molecules interact with the AQP0 tetrameric complex.
Figure 4.
Location of cross-linked lysine residues in ezrin. Cross-linked lysine residues in ezrin (PDB entry 1NI2) are highlighted in spacefill format. Lys3 (red) and Lys72 (yellow) are in subdomain F1 and Lys211 (cyan), Lys262 (green) and Lys263 (blue) are in subdomain F3.
The N246 residue of AQP0 in these identified cross-linked peptides is deamidated as is commonly seen in lens fiber cells.37 The deamidated residue D246 was the cross-linked residue in several cross-linked peptides identified which indicates that deamidation occurs before cross-linking. Note that deamidation is not required for interaction as several Glu residues were also observed to form cross-links.
Coimmunoprecipitation
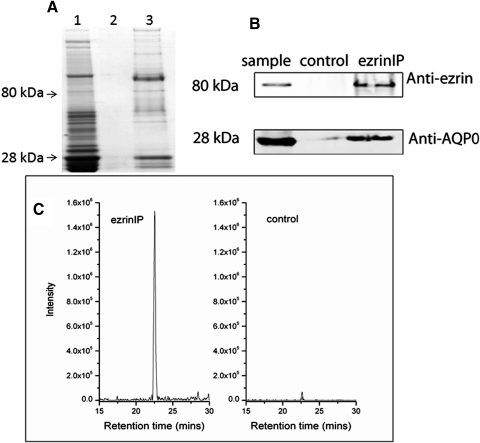
To confirm the interaction between ezrin and AQP0, coimmunoprecipitation was conducted. An ezrin antibody was added to a detergent extract of the lens WIF. Proteins that bind with the ezrin antibody were separated by SDS-PAGE. The proteins in the gel were either stained by Coomassie blue or transferred to nitrocellulose membrane for Western blotting of ezrin and AQP0. The results are shown in Figure 5. Weak bands at 80- and 28 kDa can be seen by Coomassie blue staining (Fig. 5A). Note that the strong band above 80 kDa in lane 3 is likely caused by the ezrin primary antibody. The presence of ezrin and AQP0 was confirmed by Western blotting (Fig. 5B), which indicates that the ezrin antibody precipitated both ezrin and AQP0. Ezrin was not detected in the control sample by Western blotting, but there is a very weak AQP0 signal in the control sample by Western blotting indicating a small amount of nonspecific binding of AQP0 by the protein G beads. The signal for AQP0 in the immunoprecipitated sample was much stronger than the signal in the control sample; therefore, the majority of AQP0 in the ezrin immunoprecipitated sample was related to the interaction with ezrin. The AQP0 signal in the Western blotting can be completely blocked by the AQP0 peptide that was used to produce the rabbit AQP0 antibody (data not shown).
Figure 5.
Coimmunoprecipitation of AQP0 with ezrin antibody. Lens fiber cell WIF was extracted with 25 mM tris, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 1 mM PMSF, 0.1% SDS, pH 7.4 (lane 1) and immunoprecipitated with anti-ezrin antibody (lane 3). The control sample was treated in the same way except without adding anti-ezrin antibody (lane 2). The samples were separated on SDS-PAGE and followed by Coomassie blue stain (A) and Western blotting (B). Bands at 28 and 80 kDa were detected by Coomassie blue stain in the immunoprecipitate using the ezrin antibody. The presence of ezrin and AQP0 were confirmed by Western blotting with anti-ezrin and anti-AQP0 antibodies and LC-MS/MS. Weak ezrin and AQP0 signals were also detected in the control sample, but the signals of these proteins in immunoprecipitated sample were significantly higher than in the control sample. The selected ion chromatograms of AQP0 peptide 239 to 259 (m/z 733.5) in the control sample and immunoprecipitated sample are plotted (C).
The presence of both ezrin and AQP0 in the immunoprecipitation eluate was further confirmed by mass spectrometry. The sequence coverage of ezrin and AQP0 in ezrin IP sample was 54% and 22%, respectively. Similarly, a weak signal of AQP0 was detected in the 28-kDa band of the control sample and a weak signal of ezrin was also detected in the 80-kDa band of the control sample by MS; however, their signal was dramatically lower than in the ezrin IP sample. Figure 5C shows the selected ion chromatogram of the AQP0 peptide 239 to 259 from the analysis of the ezrin IP sample. The peak area for this AQP0 peptide in the ezrin IP sample is 40 times more than the peak area seen in the control sample.
To investigate whether components of the EPPD complex are present in the ezrin IP sample, eluates from mouse IgG conjugated beads and ezrin antibody conjugated beads were run into a short stack gel and the entire stained area was cut, digested by trypsin, and analyzed by LC-MS/MS. Buffers of varying stringency were used during the IP including RIPA buffer (0.1% SDS, 0.5% deoxycholate, 1% Nonidet P-40, 150 mM NaCl, 50 mM Tris, 1mM PMSF, pH 7.5) and Tris/Triton(1%) buffer (25 mM Tris, 1 mM PMSF, 10 mM NaF, 150 mM NaCl) with or without 0.1% SDS. AQP0 and EPPD proteins periaxin and desmoyokin were detected in ezrin conjugated beads, but not in the control sample for immunoprecipitation performed in the Tris/Triton buffer with or without 0.1% SDS. Periaxin and desmoyokin were not detected in immunoprecipitations performed in the most stringent RIPA buffer; however, both ezrin and AQP0 were detected. Neither ezrin nor AQP0 were detected using the control mouse IgG conjugated beads. Periplakin was not detected under any condition.
AQP0 C-Terminal Peptide Pulldown
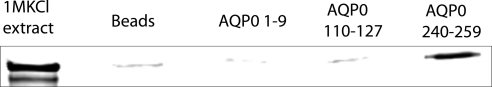
To confirm the interaction between the AQP0 C terminus with ERM protein, 1 M KCl extracted bovine lens WIF was incubated with streptavidin beads conjugated with the AQP0 C-terminal peptide(240–259). Control experiments were designed with beads alone or with beads conjugated to either an AQP0 loop peptide (residues 110–127) or to the AQP0 N terminus (residues 1–9). Proteins that bound to each bead type were eluted and resolved by SDS-PAGE and transferred to nitrocellulose membrane for Western blot analysis with an ezrin antibody. The results are shown in Figure 6, which shows a specific interaction between the AQP0 C-terminal peptide and ezrin. Weak nonspecific binding of ezrin with control beads was detected, but significantly more ezrin was bound to beads conjugated with the AQP0 C-terminal peptide.
Figure 6.
AQP0 C-terminal peptide pulldown. C-terminal AQP0 peptide (240–259) was immobilized on streptavidin beads. AQP0 loop peptide (110–127) and N-terminal peptide (1–9) containing beads and streptavidin beads alone were used as controls. KCl (1M) extract of WIF was loaded onto the beads and proteins specifically bound with beads were eluted using SDS sample buffer and Western blotted with an ezrin antibody. Lane 1, 1 M KCl extract; lane 2, beads alone; lane 3, N-terminal peptide (1–9); lane 4, loop peptide (110–127); lane 5, C-terminal peptide (240–259). Ezrin was identified as AQP0 C-terminal peptide binding partner as evidenced by significantly more ezrin bound with C-terminal peptide conjugated beads than the controls.
Discussion
ERM proteins, as plasma membrane–cytoskeleton linkers, can directly or indirectly bind with membrane proteins. Several direct membrane binding partners have been identified including CD44, CD43, PSGL-1, ICAM-1–3, and VCAM-1.38–41 The crystal structure of the radixin FERM domain bound to the cytoplasmic tail of ICAM2, CD43, or PSGL1 identified a consensus sequence, Arg/Lys/Gln-X-X-Thr-Tyr/Leu-X-X-Ala/Gly (motif-1), that binds in a groove formed by an α-helix and β-strands of subdomain F3.42 As the first identified ERM direct binding partner, CD44 lacks this sequence homology; however, it binds in the same groove in subdomain F3.43 In epithelial cells, ERM proteins indirectly interact with membrane proteins through ezrin binding phosphoprotein 50 (EBP50), a protein widely distributed in tissues, especially in those containing polarized epithelia.44,45 The crystal structure of the moesin FERM domain bound to the EBP50 C-terminal peptide indicated a different binding site on subdomain F3.42 These results show the existence of versatile ERM binding partners. In the present study, three independent experimental strategies were used to discover and confirm a direct interaction between ezrin and the C terminus of AQP0. The AQP0 C terminus does not have a consensus sequence similar to the aforementioned adhesion molecules or to EBP50; however, the cross-linking data indicate the exact sites of binding on both AQP0 and ezrin (Table 1). The cross-linking data indicate that the binding site on subdomain F3 is in the region where EBP50 is bound. Different from the other ERM binding partners reported previously, AQP0 is found to interact with both ezrin subdomains F1 and F3, which are separated three-dimensionally. The crystal structure of the radixin FERM domain with P-selectin glycoprotein ligand-1peptide (2EMT of PDB entry) also indicates the potential interaction of subdomain F1 with F3 binding partners.46 Additional studies are needed to understand this interaction; however, there are several possible explanations: (1) because the ERM proteins are present in two forms—the active form and the dormant form22—different ERM subdomain interactions with AQP0 could arise from different forms of ERM proteins; (2) the ERM proteins exist mainly in the form of dimers or higher order oligomers,47 and in the latter, the interacting subdomains F1 and F3 could be from different monomers that, when polymerized, bring subdomain F1 of one molecule close to subdomain F3 of another molecule; and (3) more than one ERM protein interacts with a single AQP0 tetramer.
The cross-linker EDC used in this study is a zero-length cross-linker, and it cross-links two reactive residues in very close proximity allowing for the detection of specific protein–protein interactions with great confidence. The AQP0 C terminus was found specifically cross-linked with relative low abundance ERM proteins and only one other, previously identified protein in the water insoluble fraction. In addition, the cross-linked residues in ERM proteins are not randomly distributed throughout the protein but are located close to one another in either the F1 or F3 subdomain. These results indicate the specificity of the cross-linking reaction. We note that the AQP0 C terminus has been reported to interact with other proteins, such as calmodulin48 and filensin.21 The region of AQP0 that interacts with calmodulin is in the juxtamembrane region,18 which is different from the more distal C-terminal region reported here for interaction with ERM proteins. Based on previous results, this distal C-terminal region of AQP0 may also be the interaction site for filensin interaction21; however, interaction with filensin and ezrin could occur in the different regions of the lens corresponding to different stages of fiber cell differentiation.
AQP0 belongs to the superfamily of aquaporins and contributes to more than 50% of the total membrane protein in the lens fiber cell.3 In addition to its function as a water channel, AQP0 has been reported to be involved in cell–cell adhesion.10–15 Previously, Straub et al.1 reported a novel cell–cell junction system containing EPPD complex, which is located on both the short and long sides of hexagonal lens fiber cells.49 It is unclear how the EPPD complex is linked to the plasma membrane, because the transmembrane protein in this complex has not been determined. The known ezrin-binding protein CD44 was not found in the EPPD complex.1 The direct interaction identified in this article suggests that AQP0 may be a candidate ezrin-binding parter in this complex. Coimmunoprecipitation results suggest that periaxin and desmoyokin coprecipitated with ezrin and AQP0; however, periplakin was not detected in our experiment by MS-based protein identification. Additional experiments are needed to confirm whether AQP0 play a role in linking EPPD complex to the cell membrane.
The ERM family is involved in many important cellular processes, including cell–cell adhesion, the maintenance of cell shape, cell motility, and membrane trafficking.25 ERM proteins have been proposed to be important in lens differentiation and functions,28 and the direct study of the function of ERM protein in the process of lens differentiation is absent; however, the ERM C-terminal binding partner actin has been identified as one of the major cytoskeletal proteins in the lens50,51 and is believed to play an important role during fiber cell differentiation and elongation. For example, lens fiber cell elongation is accompanied by increased actin stress fiber formation,52,53 and the disruption of the actin cytoskeleton impairs lens fiber cell elongation.54,55 Functional knockout of RhoGTPase, a small GTP-binding protein targeting to the actin cytoskeleton, impaired cytoskeletal organization and membrane integrity.56 Overexpression of Rho GDP dissociation inhibitor disrupted fiber cell migration, elongation, organization, and affected lens cell proliferation, differentiation, and survival, including significantly reducing AQP0 expression.57 Knockout of actin-binding protein tropomodulin 1 also disrupted membrane skeleton organization and hexagonal geometry of fiber cells.58 These results show the essential role of the actin cytoskeleton in the lens fiber cells. The interaction of AQP0 with ERM proteins suggests important roles of AQP0 during lens fiber cell elongation and differentiation. Moreover, because the AQP0 C terminus is extensively modified with age15,16 and ezrin is also degraded in the nucleus,59,60 this interaction is expected to occur in differentiating fiber cells but not in older fiber cells. How posttranslational modifications of AQP0 alter the AQP0 and ERM interaction specifically requires additional investigation.
Acknowledgments
We acknowledge the Proteomics Core of the Mass Spectrometry Research Center at Vanderbilt University.
Footnotes
Supported by National Institutes of Health Grants EY013462 (KLS) and P30EY08126 (Vanderbilt Vision Research Center).
Disclosure: Z. Wang, None; K.L. Schey, None.
References
- 1. Straub BK, Boda J, Kuhn C, et al. A novel cell-cell junction system: the cortex adhaerens mosaic of lens fiber cells. J Cell Sci. 2003;116:4985–4995 [DOI] [PubMed] [Google Scholar]
- 2. Kuwabara T, Kinoshita JH, Cogan DG. Electron microscopic study of galactose-induced cataract. Invest Ophthalmol. 1969;8:133–149 [PubMed] [Google Scholar]
- 3. Benedetti EL, Dunia I, Bentzel CJ, Vermorken AJ, Kibbelaar M, Bloemendal H. A portrait of plasma membrane specializations in eye lens epithelium and fibers. Biochim Biophys Acta. 1976;457:353–384 [DOI] [PubMed] [Google Scholar]
- 4. Francis P, Chung JJ, Yasui M, et al. Functional impairment of lens aquaporin in two families with dominantly inherited cataracts. Hum Mol Genet. 2000;9:2329–2334 [DOI] [PubMed] [Google Scholar]
- 5. Shiels A, Mackay D, Bassnett S, Al-Ghoul K, Kuszak J. Disruption of lens fiber cell architecture in mice expressing a chimeric AQP0-LTR protein. FASEB J. 2000;14:2207–2212 [DOI] [PubMed] [Google Scholar]
- 6. Bateman JB, Johannes M, Flodman P, et al. A new locus for autosomal dominant cataract on chromosome 12q13. Invest Ophthalmol Vis Sci. 2000;41:2665–2670 [PubMed] [Google Scholar]
- 7. Berry V, Francis P, Kaushal S, Moore A, Bhattacharya S. Missense mutations in MIP underlie autosomal dominant ‘polymorphic’ and lamellar cataracts linked to 12q. Nat Genet. 2000;25:15–17 [DOI] [PubMed] [Google Scholar]
- 8. Kushmerick C, Rice SJ, Baldo GJ, Haspel HC, Mathias RT. Ion, water and neutral solute transport in Xenopus oocytes expressing frog lens MIP. Exp Eye Res. 1995;61:351–362 [DOI] [PubMed] [Google Scholar]
- 9. Chandy G, Zampighi GA, Kreman M, Hall JE. Comparison of the water transporting properties of MIP and AQP1. J Membr Biol. 1997;159:29–39 [DOI] [PubMed] [Google Scholar]
- 10. Zampighi GA, Hall JE, Ehring GR, Simon SA. The structural organization and protein composition of lens fiber junctions. J Cell Biol. 1989;108:2255–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dunia I, Manenti S, Rousselet A, Benedetti EL. Electron microscopic observations of reconstituted proteoliposomes with the purified major intrinsic membrane protein of eye lens fibers. J Cell Biol. 1987;105:1679–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Michea LF, de la Fuente M, Lagos N. Lens major intrinsic protein (MIP) promotes adhesion when reconstituted into large unilamellar liposomes. Biochemistry. 1994;33:7663–7669 [DOI] [PubMed] [Google Scholar]
- 13. Gonen T, Cheng Y, Sliz P, et al. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kumari SS, Varadaraj K. Intact AQP0 performs cell-to-cell adhesion. Biochem Biophys Res Commun. 2009;390:1034–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Varadaraj K, Kumari SS, Mathias RT. Transgenic expression of AQP1 in the fiber cells of AQP0 knockout mouse: effects on lens transparency. Exp Eye Res. 2010;91:393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schey KL, Fowler JG, Schwartz JC, Busman M, Dillon J, Crouch RK. Complete map and identification of the phosphorylation site of bovine lens major intrinsic protein. Invest Ophthalmol Vis Sci. 1997;38:2508–2515 [PubMed] [Google Scholar]
- 17. Schey KL, Little M, Fowler JG, Crouch RK. Characterization of human lens major intrinsic protein structure. Invest Ophthalmol Vis Sci. 2000;41:175–182 [PubMed] [Google Scholar]
- 18. Rose KM, Wang Z, Magrath GN, Hazard ES, Hildebrandt JD, Schey KL. Aquaporin 0-calmodulin interaction and the effect of aquaporin 0 phosphorylation. Biochemistry. 2008;47:339–347 [DOI] [PubMed] [Google Scholar]
- 19. Kalman K, Németh-Cahalan KL, Froger A, Hall JE. Phosphorylation determines the calmodulin-mediated Ca2+ response and water permeability of AQP0. J Biol Chem. 2008;283:21278–21283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Varadaraj K, Kumari S, Shiels A, Mathias RT. Regulation of aquaporin water permeability in the lens. Invest Ophthalmol Vis Sci. 2005;46:1393–1402 [DOI] [PubMed] [Google Scholar]
- 21. Lindsey Rose KM, Gourdie RG, Prescott AR, Quinlan RA, Crouch RK, Schey KL. The C terminus of lens aquaporin 0 interacts with the cytoskeletal proteins filensin and CP49. Invest Ophthalmol Vis Sci. 2006;47:1562–1570 [DOI] [PubMed] [Google Scholar]
- 22. Bretscher A, Reczek D, Berryman M. Ezrin: a protein requiring conformational activation to link microfilaments to the plasma membrane in the assembly of cell surface structures. J Cell Sci. 1997;110:3011–3018 [DOI] [PubMed] [Google Scholar]
- 23. Tsukita S, Yonemura S, Tsukita S. ERM proteins: head-to-tail regulation of actin-plasma membrane interaction. Trends Biochem Sci. 1997;22:53–58 [DOI] [PubMed] [Google Scholar]
- 24. Vaheri A, Carpén O, Heiska L, et al. The ezrin protein family: membrane-cytoskeleton interactions and disease associations. Curr Opin Cell Biol. 1997;9:659–666 [DOI] [PubMed] [Google Scholar]
- 25. Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11:276–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chishti AH, Kim AC, Marfatia SM, et al. The FERM domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem Sci. 1998;23:281–282 [DOI] [PubMed] [Google Scholar]
- 27. Aster JC, Brewer GJ, Hanash SM, Maisel H. Band 4.1-like proteins of the bovine lens. Effects of differentiation, distribution and extraction characteristics. Biochem J. 1984;224:609–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bagchi M, Katar M, Lo WK, Yost R, Hill C, Maisel H. ERM proteins of the lens. J Cell Biochem. 2004;92:626–630 [DOI] [PubMed] [Google Scholar]
- 29. Rao PV, Ho T, Skiba NP, Maddala R. Characterization of lens fiber cell triton insoluble fraction reveals ERM (ezrin, radixin, moesin) proteins as major cytoskeletal-associated proteins. Biochem Biophys Res Commun. 2008;368:508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247 [DOI] [PubMed] [Google Scholar]
- 31. Rinner O, Seebacher J, Walzthoeni T, et al. Identification of cross-linked peptides from large sequence databases. Nat Methods. 2008;5:315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ma ZQ, Dasari S, Chambers MC, et al. IDPicker 2.0: improved protein assembly with high discrimination peptide identification filtering. J Proteome Res. 2009;8:3872–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schilling B, Row RH, Gibson BW, Guo X, Young MM. MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J Am Soc Mass Spectrom. 2003;14:834–850 [DOI] [PubMed] [Google Scholar]
- 34. Fiévet B, Louvard D, Arpin M. ERM proteins in epithelial cell organization and functions. Biochim Biophys Acta. 2007;1773:653–660 [DOI] [PubMed] [Google Scholar]
- 35. Wang Z, Obidike JE, Schey KL. Posttranslational modifications of the bovine lens beaded filament proteins filensin and CP49. Invest Ophthalmol Vis Sci. 2010;51:1565–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smith WJ, Nassar N, Bretscher A, Cerione RA, Karplus PA. Structure of the active N-terminal domain of Ezrin. Conformational and mobility changes identify keystone interactions. J Biol Chem. 2003;278:4949–4956 [DOI] [PubMed] [Google Scholar]
- 37. Ball LE, Garland DL, Crouch RK, Schey KL. Post-translational modifications of aquaporin 0 (AQP0) in the normal human lens: spatial and temporal occurrence. Biochemistry. 2004;43:9856–9865 [DOI] [PubMed] [Google Scholar]
- 38. Tsukita S, Oishi K, Sato N, Sagara J, Kawai A, Tsukita S. ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J Cell Biol. 1994;126:391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Serrador JM, Alonso-Lebrero JL, del Pozo MA, et al. Moesin interacts with the cytoplasmic region of intercellular adhesion molecule-3 and is redistributed to the uropod of T lymphocytes during cell polarization. J Cell Biol. 1997;138:1409–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heiska L, Alfthan K, Grönholm M, Vilja P, Vaheri A, Carpén O. Association of ezrin with intercellular adhesion molecule-1 and −2 (ICAM-1 and ICAM-2). Regulation by phosphatidylinositol 4, 5-bisphosphate. J Biol Chem. 1998;273:21893–21900 [DOI] [PubMed] [Google Scholar]
- 41. Yonemura S, Hirao M, Doi Y, Takahashi N, Kondo T, Tsukita S. Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J Cell Biol. 1998;140:885–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Terawaki S, Maesaki R, Hakoshima T. Structural basis for NHERF recognition by ERM proteins. Structure. 2006;14:777–789 [DOI] [PubMed] [Google Scholar]
- 43. Mori T, Kitano K, Terawaki S, Maesaki R, Fukami Y, Hakoshima T. Structural basis for CD44 recognition by ERM proteins. J Biol Chem. 2008;283:29602–29612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morales FC, Takahashi Y, Kreimann EL, Georgescu MM. Ezrin-radixin-moesin (ERM)-binding phosphoprotein 50 organizes ERM proteins at the apical membrane of polarized epithelia. Proc Natl Acad Sci U S A. 2004;101:17705–17710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Finnerty CM, Chambers D, Ingraffea J, Faber HR, Karplus PA, Bretscher A. The EBP50-moesin interaction involves a binding site regulated by direct masking on the FERM domain. J Cell Sci. 2004;117:1547–1552 [DOI] [PubMed] [Google Scholar]
- 46. Takai Y, Kitano K, Terawaki S, Maesaki R, Hakoshima T. Structural basis of PSGL-1 binding to ERM proteins. Genes Cells. 2007;12:1329–1338 [DOI] [PubMed] [Google Scholar]
- 47. Berryman M, Gary R, Bretscher A. Ezrin oligomers are major cytoskeletal components of placental microvilli: a proposal for their involvement in cortical morphogenesis. J Cell Biol. 1995;131:1231–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Németh-Cahalan KL, Hall JE. pH and calcium regulate the water permeability of aquaporin 0. J Biol Chem. 2000;275:6777–6782 [DOI] [PubMed] [Google Scholar]
- 49. Song S, Landsbury A, Dahm R, Liu Y, Zhang Q, Quinlan RA. Functions of the intermediate filament cytoskeleton in the eye lens. J Clin Invest. 2009;119:1837–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ireland M, Lieska N, Maisel H. Lens actin: purification and localization. Exp Eye Res. 1983;37:393–408 [DOI] [PubMed] [Google Scholar]
- 51. Kibbelaar MA, Selten-Versteegen AM, Dunia I, Benedetti EL, Bloemendal H. Actin in mammalian lens. Eur J Biochem. 1979;95:543–549 [DOI] [PubMed] [Google Scholar]
- 52. Courtois Y, Arruti C, Barritault D, Tassin J, Olivié M, Hughes RC. Modulation of the shape of epithelial lens cells in vitro directed by a retinal extract factor. A model of interconversions and the role of actin filaments and fibronectin. Differentiation. 1981;18:11–27 [DOI] [PubMed] [Google Scholar]
- 53. Ramaekers F, Jap P, Mungyer G, Bloemendal H. Microfilament assembly during lens cell elongation in vitro. Curr Eye Res. 1982;2:169–181 [DOI] [PubMed] [Google Scholar]
- 54. Beebe DC, Cerrelli S. Cytochalasin prevents cell elongation and increases potassium efflux from embryonic lens epithelial cells: implications for the mechanism of lens fiber cell elongation. Lens Eye Toxic Res. 1989;6:589–601 [PubMed] [Google Scholar]
- 55. Mousa GY, Trevithick JP. Differentiation of rat lens epithelial cells in tissue culture. II. Effect of cytochalasin B and D on actin organization and differentiation. Dev Biol. 1977;60:14–25 [DOI] [PubMed] [Google Scholar]
- 56. Maddala R, Deng PF, Costello JM, Wawrousek EF, Zigler JS, Rao VP, Impaired cytoskeletal organization and membrane integrity in lens fibers of a Rho GTPase functional knockout transgenic mouse. Lab Invest. 2004;84:679–692 [DOI] [PubMed] [Google Scholar]
- 57. Maddala R, Reneker LW, Pendurthi B, Rao PV. Rho GDP dissociation inhibitor-mediated disruption of Rho GTPase activity impairs lens fiber cell migration, elongation and survival. Dev Biol. 2008;315:217–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nowak RB, Fischer RS, Zoltoski RK, Kuszak JR, Fowler VM. Tropomodulin1 is required for membrane skeleton organization and hexagonal geometry of fiber cells in the mouse lens. J Cell Biol. 2009;186:915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Watanabe M, Kobayashi H, Yao R, Maisel H. Adhesion and junction molecules in embryonic and adult lens cell differentiation. Acta Ophthalmol Suppl. 1992;205:46–52 [DOI] [PubMed] [Google Scholar]
- 60. Wang Z, Han J, Schey KL. Spatial differences in an integral membrane proteome detected in laser capture microdissected samples. J Proteome Res. 2008;7:2696–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]