Figure 3 .
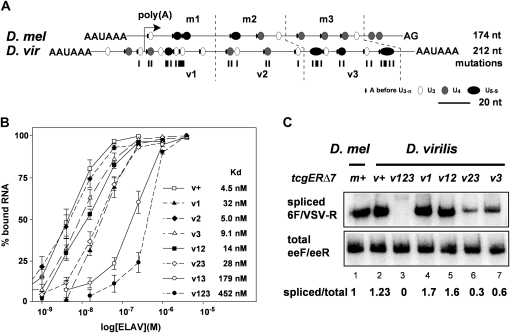
Requirement of multiple short poly(U) motives for ewg intron 6 splicing is conserved in D. virilis. (A) Schematic of the ELAV-binding site at pA2 from D. melanogaster and at virpA from D. virilis centered at the cleavage site. The ELAV-binding site is divided into three parts for each species: m1–3 for D. melanogaster and v1–3 for D. virilis. Poly(U) motifs are indicated as circles according to their length, starting with three Us, and A in front of the poly(U) motifs is indicated as a line. Mutations, mostly U-to-C substitutions, introduced into the D. virilis ELAV-binding site are indicated below as lines. (B) Graphic representation of EMSA data using recombinant D. melanogaster ELAV and virpA substrate RNAs containing mutations in the parts depicted in A as means with standard error from three to five experiments. EMSAs were done by incubating uniformly 32P-labeled RNAs (100 pM) with recombinant ELAV in five to six concentrations over the binding range (1, 3.9, 15.6, and 62.5 nM and 0.25, 1, and 4 µM) separated on 4% native polyacrylamide gels. The percentage of bound RNA (input RNA-unbound RNA/input RNA × 100) is plotted against the concentration of recombinant ELAV (in morgans) presented as log. (C) Semiquantitative RT-PCR of intron 6 splicing using 32P-labeled forward primers from tcgER∆7m and v transgenes containing mutated parts of the D. virilis ELAV-binding site as depicted in A in third instar larval brains using primers 6F and VSV-R (cycle 26) compared to total expression levels of tcgER∆7 (primers eeF and eeR, cycle 28) analyzed on 8% polyacrylamide gels. Quantification of three experiments is shown at the bottom.