Figure 3.
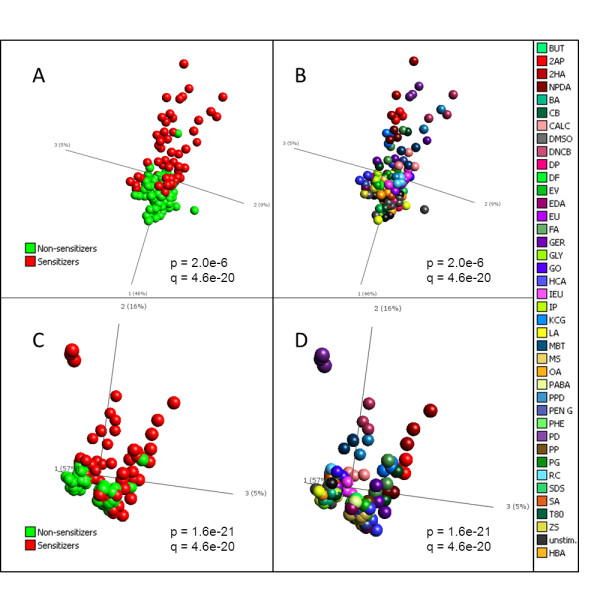
Principal component analysis of transcripts differentially expressed after chemical stimulation. mRNA levels in MUTZ-3 cells stimulated for 24 h with 20 sensitizing and 20 non-sensitizing chemicals were assessed with transcriptomics, using Affymetrix Human Gene 1.0 ST arrays. Structures and similarities in the gene expression data set were investigated, using principal component analysis (PCA) in the software Qlucore. A) PCA of genes differentially expressed in cells stimulated with sensitizing (red) versus non-sensitizing (green) chemicals (1010 genes identified with one-way ANOVA). B) PCA of genes differentially expressed in cells stimulated with sensitizing versus non-sensitizing chemicals (1010 genes), but now samples are colored by the compound used for stimulation. C) PCA of genes differentially expressed when comparing the different stimulations with 2-way ANOVA (1137 genes). Samples are colored according to sensitizing (red) and non-sensitizing (green) chemicals. D) PCA of genes differentially expressed when comparing the different stimulations with 2-way ANOVA (1137 genes), but now samples are colored by the compound used for stimulation. P, p-value from ANOVA. Q, p-value corrected for multiple hypothesis testing.
