Fig. 1.
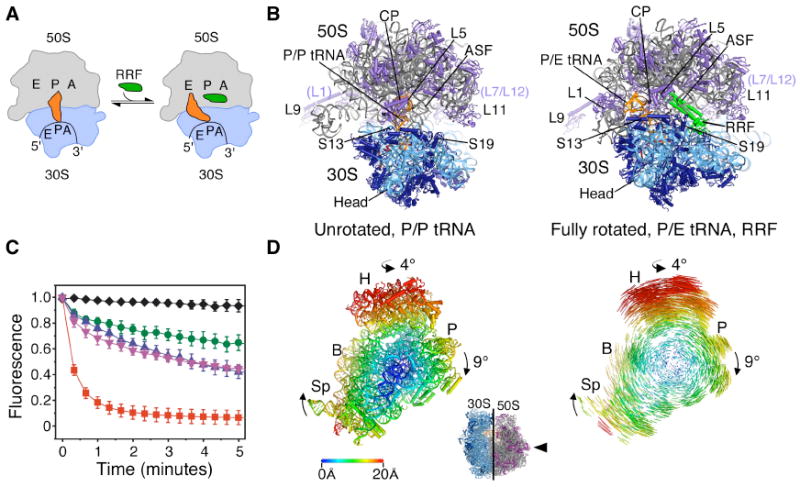
Ribosome recycling in bacteria and organelles. (A) Steps of ribosome recycling. After termination, ribosomes with deacylated tRNA in the P site undergo a structural rearrangement to a fully rotated state in which tRNA adopts a P/E hybrid state of binding and RRF is bound in the 50S P site. EF-G then catalyzes subunit dissociation (not shown). (B) Global views of the ribosome in a post-termination state (L) and intermediate state of recycling (R). The small subunit rRNA and proteins are colored light and dark blue, respectively, with the large subunit rRNA and proteins colored grey and magenta, respectively. Bound tRNA (orange), mRNA (dark red), and RRF (green) are also shown. (C) The dependence of subunit release on RRF, EF-G and GTP under crystallographic buffer conditions. Release was monitored by the loss of Cy5-labeled L1 fluorescence in 50S subunits from surface-immobilized ribosome complexes carrying Cy3-labeled tRNAPhe in the P site. Complexes imaged in the absence of factors (black diamonds) or in the presence of 10μM RRF (green circles), 20 μM EF-G/2mM GTP (pink inverted triangles), 10 μM RRF/20 μM EF-G/2mM GDPNP (blue triangles) or 10 μM RRF/20 μM EF-G/2mM GTP (red squares). Data reflect the mean +/- SD of normalized Cy5 fluorescence intensity as a function of time from three experimental replicates. (D) Conformational changes in the 70S ribosome during ratcheting. View of the 30S subunit from the perspective of the 50S subunit (inset). Shifts between equivalent RNA phosphorus atoms and protein Cα atoms in the unrotated (R0) and fully rotated (RF) states are color coded as indicated by the scale. Ribosomes were superimposed using the 50S subunit as the frame of reference (37). Difference vectors between equivalent phosphorus or Cα atoms of the 30S subunits in the unrotated and fully rotated ribosome structures are shown on the right.
