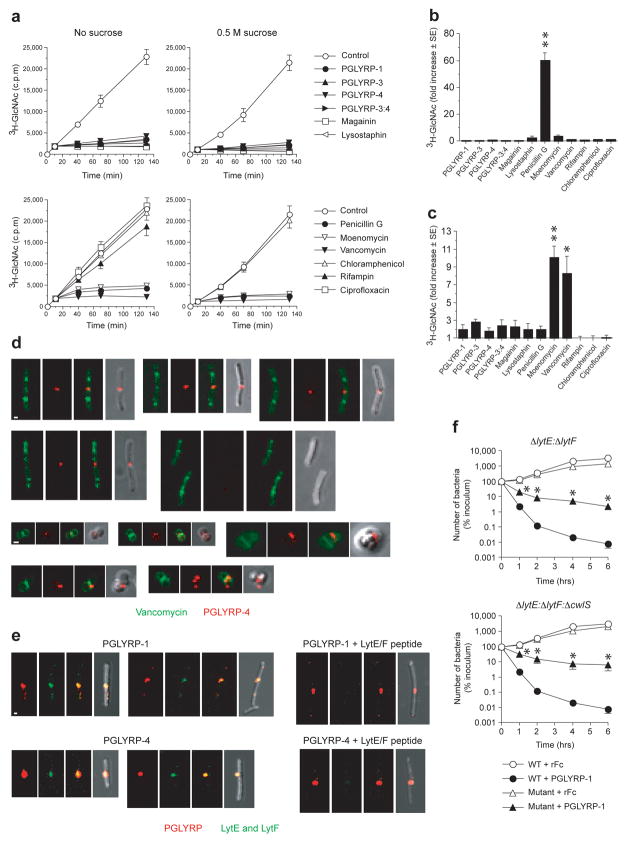
Figure 1. PGRPs inhibit intracellular step in peptidoglycan synthesis and localize to the newly formed cell separation site.
(a) PGRPs inhibit peptidoglycan synthesis in S. aureus measured by incorporation of 3H-GlcNAc into insoluble peptidoglycan; 0.5 M sucrose prevents osmotic lysis; means ± SEM of 3 – 4 experiments. (b) PGRPs do not inhibit transpeptidation in S. aureus measured by incorporation of 3H-GlcNAc into secreted uncrosslinked polymeric peptidoglycan. (c) PGRPs do not inhibit transglycosylation in S. aureus measured as accumulation of 3H-GlcNAc-lipid II in cell membrane; means ± SEM of 3–4 (b) or 4–5 (c) experiments; *, P<0.03; **, P<0.0002 (t-test). (d) PGRPs localize to the newly formed cell separation site and not to the sites of peptidoglycan synthesis in B. subtilis (top rows) or S. aureus (bottom rows); different stages of cell division shown; each panel from left to right shows green (BODIPY-FL-vancomycin), red (Alexa Fluor 594-PGLYRP-4), overlay red and green, and overlay of red and dark field view. (e) PGLYRP-1 or PGLYRP-4 (red) co-localize with LytE and LytF (green) in B. subtilis and LytE/LytF peptide inhibits detection LytE and LytF; each panel from left to right shows red (PGRP), green (LytE and LytF), overlay red and green (yellow), and overlay of red and green and dark field view; bar = 1 μm. (f) LytE- and LytF-generated cell separation sites are required for efficient killing of B. subtilis by PGRPs, shown by lower killing of ΔlytE:ΔlytF or ΔlytE: ΔlytF: ΔcwlS mutants than of WT bacteria by PGLYRP-1; means ± SEM of 3 experiments; *, P<0.05 versus WT (t-test).