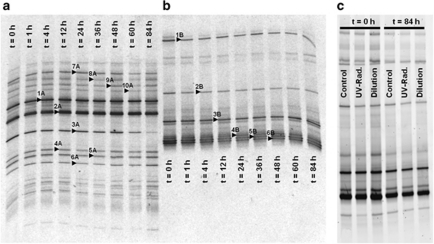
Figure 4.
DGGE profiles from archaeal (a) and bacterial (b) 16S rRNA gene amplification products in the samples used as controls. Each lane corresponds to different sampling times in the experiment (in hours). In (c), comparison among the bacterial assemblages in the control, diluted and UV-radiated samples in two different sampling times. Sequences 1, 2 and 10, in (a), were related to the species Hqr. walsbyi. The rest of the bands in (a) were associated to uncultured Halobacteriaceae environmental clones. Sequences from band 3 were related to the species S. ruber. The rest of the sequences in (b) were associated to uncultured Salinibacter (bands 2, 4-6) and Bacteroidetes (band 1) clones.